Genome-wide CRISPR-KO Screen Uncovers mTORC1-Mediated Gsk3 Regulation in Naive Pluripotency Maintenance and Dissolution
- PMID: 29996108
- PMCID: PMC6057492
- DOI: 10.1016/j.celrep.2018.06.027
Genome-wide CRISPR-KO Screen Uncovers mTORC1-Mediated Gsk3 Regulation in Naive Pluripotency Maintenance and Dissolution
Abstract
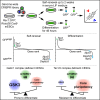
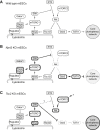
The genetic basis of naive pluripotency maintenance and loss is a central question in embryonic stem cell biology. Here, we deploy CRISPR-knockout-based screens in mouse embryonic stem cells to interrogate this question through a genome-wide, non-biased approach using the Rex1GFP reporter as a phenotypic readout. This highly sensitive and efficient method identified genes in diverse biological processes and pathways. We uncovered a key role for negative regulators of mTORC1 in maintenance and exit from naive pluripotency and provided an integrated account of how mTORC1 activity influences naive pluripotency through Gsk3. Our study therefore reinforces Gsk3 as the central node and provides a comprehensive, data-rich resource that will improve our understanding of mechanisms regulating pluripotency and stimulate avenues for further mechanistic studies.
Keywords: Akt; CRISPR; GATOR1; Nprl2; Tsc2; exit from pluripotency; mTORC1; mTORC2; naive pluripotency; screening.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Intracellular Ca2+ Homeostasis and Nuclear Export Mediate Exit from Naive Pluripotency.Cell Stem Cell. 2019 Aug 1;25(2):210-224.e6. doi: 10.1016/j.stem.2019.04.015. Epub 2019 May 16. Cell Stem Cell. 2019. PMID: 31104942 Free PMC article.
-
A Genome-Wide CRISPR Screen Identifies Factors Regulating Pluripotency Exit in Mouse Embryonic Stem Cells.Cells. 2022 Jul 25;11(15):2289. doi: 10.3390/cells11152289. Cells. 2022. PMID: 35892587 Free PMC article.
-
Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation.Nat Genet. 2019 Jun;51(6):999-1010. doi: 10.1038/s41588-019-0408-9. Epub 2019 May 20. Nat Genet. 2019. PMID: 31110351 Free PMC article.
-
Decoding pluripotency: Genetic screens to interrogate the acquisition, maintenance, and exit of pluripotency.Wiley Interdiscip Rev Syst Biol Med. 2020 Jan;12(1):e1464. doi: 10.1002/wsbm.1464. Epub 2019 Aug 13. Wiley Interdiscip Rev Syst Biol Med. 2020. PMID: 31407519 Free PMC article. Review.
-
β-Catenin in pluripotency: adhering to self-renewal or Wnting to differentiate?Int Rev Cell Mol Biol. 2014;312:53-78. doi: 10.1016/B978-0-12-800178-3.00002-6. Int Rev Cell Mol Biol. 2014. PMID: 25262238 Review.
Cited by
-
Using CRISPR to understand and manipulate gene regulation.Development. 2021 May 1;148(9):dev182667. doi: 10.1242/dev.182667. Epub 2021 Apr 29. Development. 2021. PMID: 33913466 Free PMC article. Review.
-
The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis.Cell Stem Cell. 2019 Nov 7;25(5):622-638.e13. doi: 10.1016/j.stem.2019.08.018. Epub 2019 Oct 3. Cell Stem Cell. 2019. PMID: 31588046 Free PMC article.
-
Intracellular Ca2+ Homeostasis and Nuclear Export Mediate Exit from Naive Pluripotency.Cell Stem Cell. 2019 Aug 1;25(2):210-224.e6. doi: 10.1016/j.stem.2019.04.015. Epub 2019 May 16. Cell Stem Cell. 2019. PMID: 31104942 Free PMC article.
-
Screening Genes Promoting Exit from Naive Pluripotency Based on Genome-Scale CRISPR-Cas9 Knockout.Stem Cells Int. 2020 Feb 3;2020:8483035. doi: 10.1155/2020/8483035. eCollection 2020. Stem Cells Int. 2020. PMID: 32089710 Free PMC article.
-
Electrical devices for visual restoration.Surv Ophthalmol. 2022 May-Jun;67(3):793-800. doi: 10.1016/j.survophthal.2021.08.008. Epub 2021 Sep 4. Surv Ophthalmol. 2022. PMID: 34487742 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

