Structures of human Patched and its complex with native palmitoylated sonic hedgehog
- PMID: 29995851
- PMCID: PMC6341490
- DOI: 10.1038/s41586-018-0308-7
Structures of human Patched and its complex with native palmitoylated sonic hedgehog
Abstract
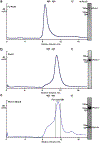
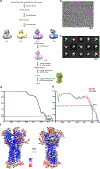
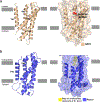
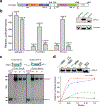
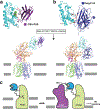
Hedgehog (HH) signalling governs embryogenesis and adult tissue homeostasis in mammals and other multicellular organisms1-3. Whereas deficient HH signalling leads to birth defects, unrestrained HH signalling is implicated in human cancers2,4-6. N-terminally palmitoylated HH releases the repression of Patched to the oncoprotein smoothened (SMO); however, the mechanism by which HH recognizes Patched is unclear. Here we report cryo-electron microscopy structures of human patched 1 (PTCH1) alone and in complex with the N-terminal domain of 'native' sonic hedgehog (native SHH-N has both a C-terminal cholesterol and an N-terminal fatty-acid modification), at resolutions of 3.5 Å and 3.8 Å, respectively. The structure of PTCH1 has internal two-fold pseudosymmetry in the transmembrane core, which features a sterol-sensing domain and two homologous extracellular domains, resembling the architecture of Niemann-Pick C1 (NPC1) protein7. The palmitoylated N terminus of SHH-N inserts into a cavity between the extracellular domains of PTCH1 and dominates the PTCH1-SHH-N interface, which is distinct from that reported for SHH-N co-receptors8. Our biochemical assays show that SHH-N may use another interface, one that is required for its co-receptor binding, to recruit PTCH1 in the absence of a covalently attached palmitate. Our work provides atomic insights into the recognition of the N-terminal domain of HH (HH-N) by PTCH1, offers a structural basis for cooperative binding of HH-N to various receptors and serves as a molecular framework for HH signalling and its malfunction in disease.
Conflict of interest statement
Figures



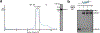
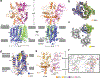
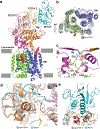
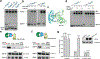
Similar articles
-
Mechanistic Insights into the Generation and Transduction of Hedgehog Signaling.Trends Biochem Sci. 2020 May;45(5):397-410. doi: 10.1016/j.tibs.2020.01.006. Epub 2020 Feb 17. Trends Biochem Sci. 2020. PMID: 32311334 Free PMC article. Review.
-
Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex.Science. 2018 Oct 5;362(6410):eaas8843. doi: 10.1126/science.aas8843. Epub 2018 Aug 23. Science. 2018. PMID: 30139912 Free PMC article.
-
Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.Nat Commun. 2019 May 24;10(1):2320. doi: 10.1038/s41467-019-10234-9. Nat Commun. 2019. PMID: 31127104 Free PMC article.
-
Structural basis of sterol recognition by human hedgehog receptor PTCH1.Sci Adv. 2019 Sep 18;5(9):eaaw6490. doi: 10.1126/sciadv.aaw6490. eCollection 2019 Sep. Sci Adv. 2019. PMID: 31555730 Free PMC article.
-
Structures of vertebrate Patched and Smoothened reveal intimate links between cholesterol and Hedgehog signalling.Curr Opin Struct Biol. 2019 Aug;57:204-214. doi: 10.1016/j.sbi.2019.05.015. Epub 2019 Jun 24. Curr Opin Struct Biol. 2019. PMID: 31247512 Free PMC article. Review.
Cited by
-
CNPY4 inhibits the Hedgehog pathway by modulating membrane sterol lipids.Nat Commun. 2022 May 3;13(1):2407. doi: 10.1038/s41467-022-30186-x. Nat Commun. 2022. PMID: 35504891 Free PMC article.
-
Regulation of Hedgehog Signal Transduction by Ubiquitination and Deubiquitination.Int J Mol Sci. 2021 Dec 11;22(24):13338. doi: 10.3390/ijms222413338. Int J Mol Sci. 2021. PMID: 34948134 Free PMC article. Review.
-
Mechanistic Insights into the Generation and Transduction of Hedgehog Signaling.Trends Biochem Sci. 2020 May;45(5):397-410. doi: 10.1016/j.tibs.2020.01.006. Epub 2020 Feb 17. Trends Biochem Sci. 2020. PMID: 32311334 Free PMC article. Review.
-
NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes.J Biol Chem. 2019 Feb 1;294(5):1706-1709. doi: 10.1074/jbc.TM118.004165. J Biol Chem. 2019. PMID: 30710017 Free PMC article. Review.
-
Hedgehog-GLI mediated control of renal formation and malformation.Front Nephrol. 2023 Apr 20;3:1176347. doi: 10.3389/fneph.2023.1176347. eCollection 2023. Front Nephrol. 2023. PMID: 37675356 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

