Genetic determinants of co-accessible chromatin regions in activated T cells across humans
- PMID: 29988122
- PMCID: PMC6097927
- DOI: 10.1038/s41588-018-0156-2
Genetic determinants of co-accessible chromatin regions in activated T cells across humans
Abstract
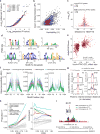
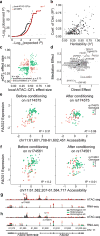
Over 90% of genetic variants associated with complex human traits map to non-coding regions, but little is understood about how they modulate gene regulation in health and disease. One possible mechanism is that genetic variants affect the activity of one or more cis-regulatory elements leading to gene expression variation in specific cell types. To identify such cases, we analyzed ATAC-seq and RNA-seq profiles from stimulated primary CD4+ T cells in up to 105 healthy donors. We found that regions of accessible chromatin (ATAC-peaks) are co-accessible at kilobase and megabase resolution, consistent with the three-dimensional chromatin organization measured by in situ Hi-C in T cells. Fifteen percent of genetic variants located within ATAC-peaks affected the accessibility of the corresponding peak (local-ATAC-QTLs). Local-ATAC-QTLs have the largest effects on co-accessible peaks, are associated with gene expression and are enriched for autoimmune disease variants. Our results provide insights into how natural genetic variants modulate cis-regulatory elements, in isolation or in concert, to influence gene expression.
Conflict of interest statement
Figures




Similar articles
-
Epigenetic and transcriptional dysregulation in CD4+ T cells in patients with atopic dermatitis.PLoS Genet. 2022 May 16;18(5):e1009973. doi: 10.1371/journal.pgen.1009973. eCollection 2022 May. PLoS Genet. 2022. PMID: 35576187 Free PMC article.
-
Dynamic regulatory elements in single-cell multimodal data implicate key immune cell states enriched for autoimmune disease heritability.Nat Genet. 2023 Dec;55(12):2200-2210. doi: 10.1038/s41588-023-01577-7. Epub 2023 Nov 30. Nat Genet. 2023. PMID: 38036783 Free PMC article.
-
Chromatin accessibility and gene expression during adipocyte differentiation identify context-dependent effects at cardiometabolic GWAS loci.PLoS Genet. 2021 Oct 26;17(10):e1009865. doi: 10.1371/journal.pgen.1009865. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34699533 Free PMC article.
-
Interrogating the Accessible Chromatin Landscape of Eukaryote Genomes Using ATAC-seq.Methods Mol Biol. 2021;2243:183-226. doi: 10.1007/978-1-0716-1103-6_10. Methods Mol Biol. 2021. PMID: 33606259 Review.
-
Detect accessible chromatin using ATAC-sequencing, from principle to applications.Hereditas. 2019 Aug 15;156:29. doi: 10.1186/s41065-019-0105-9. eCollection 2019. Hereditas. 2019. PMID: 31427911 Free PMC article. Review.
Cited by
-
Statistical methods for SNP heritability estimation and partition: A review.Comput Struct Biotechnol J. 2020 Jun 18;18:1557-1568. doi: 10.1016/j.csbj.2020.06.011. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32637052 Free PMC article. Review.
-
Human genetic variation determines 24-hour rhythmic gene expression and disease risk.Res Sq [Preprint]. 2024 Aug 5:rs.3.rs-4790200. doi: 10.21203/rs.3.rs-4790200/v1. Res Sq. 2024. PMID: 39149455 Free PMC article. Preprint.
-
Genetic analyses support the contribution of mRNA N6-methyladenosine (m6A) modification to human disease heritability.Nat Genet. 2020 Sep;52(9):939-949. doi: 10.1038/s41588-020-0644-z. Epub 2020 Jun 29. Nat Genet. 2020. PMID: 32601472 Free PMC article.
-
Prioritization of autoimmune disease-associated genetic variants that perturb regulatory element activity in T cells.Nat Genet. 2022 May;54(5):603-612. doi: 10.1038/s41588-022-01056-5. Epub 2022 May 5. Nat Genet. 2022. PMID: 35513721 Free PMC article.
-
Leveraging Single-Cell Populations to Uncover the Genetic Basis of Complex Traits.Annu Rev Genet. 2023 Nov 27;57:297-319. doi: 10.1146/annurev-genet-022123-110824. Epub 2023 Aug 10. Annu Rev Genet. 2023. PMID: 37562412 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

