A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity
- PMID: 29937225
- PMCID: PMC6046266
- DOI: 10.1016/j.cell.2018.05.043
A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity
Abstract
Transposable elements represent nearly half of mammalian genomes and are generally described as parasites, or "junk DNA." The LINE1 retrotransposon is the most abundant class and is thought to be deleterious for cells, yet it is paradoxically highly expressed during early development. Here, we report that LINE1 plays essential roles in mouse embryonic stem cells (ESCs) and pre-implantation embryos. In ESCs, LINE1 acts as a nuclear RNA scaffold that recruits Nucleolin and Kap1/Trim28 to repress Dux, the master activator of a transcriptional program specific to the 2-cell embryo. In parallel, LINE1 RNA mediates binding of Nucleolin and Kap1 to rDNA, promoting rRNA synthesis and ESC self-renewal. In embryos, LINE1 RNA is required for Dux silencing, synthesis of rRNA, and exit from the 2-cell stage. The results reveal an essential partnership between LINE1 RNA, Nucleolin, Kap1, and peri-nucleolar chromatin in the regulation of transcription, developmental potency, and ESC self-renewal.
Keywords: 2-cell stage; Dux; ESCs; Kap1; LINE1; MERVL; Nucleolin; hypertranscription; rRNA; retrotransposons.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
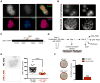
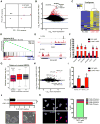
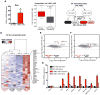
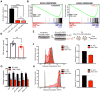
Comment in
-
A lncRNA-like Role for LINE1s in Development.Dev Cell. 2018 Jul 16;46(2):132-134. doi: 10.1016/j.devcel.2018.06.022. Dev Cell. 2018. PMID: 30016617 Free PMC article.
Similar articles
-
rRNA biogenesis regulates mouse 2C-like state by 3D structure reorganization of peri-nucleolar heterochromatin.Nat Commun. 2021 Nov 9;12(1):6365. doi: 10.1038/s41467-021-26576-2. Nat Commun. 2021. PMID: 34753899 Free PMC article.
-
Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos.Protein Cell. 2021 Jun;12(6):455-474. doi: 10.1007/s13238-021-00837-8. Epub 2021 Apr 22. Protein Cell. 2021. PMID: 33886094 Free PMC article.
-
Nucleolin maintains embryonic stem cell self-renewal by suppression of p53 protein-dependent pathway.J Biol Chem. 2011 Dec 16;286(50):43370-82. doi: 10.1074/jbc.M111.225185. Epub 2011 Oct 19. J Biol Chem. 2011. PMID: 22013067 Free PMC article.
-
Nucleolin: a multifunctional major nucleolar phosphoprotein.Crit Rev Biochem Mol Biol. 1998;33(6):407-36. doi: 10.1080/10409239891204260. Crit Rev Biochem Mol Biol. 1998. PMID: 9918513 Review.
-
Nucleolin: dual roles in rDNA chromatin transcription.Gene. 2015 Feb 1;556(1):7-12. doi: 10.1016/j.gene.2014.09.023. Epub 2014 Sep 16. Gene. 2015. PMID: 25225127 Review.
Cited by
-
MPP8 is essential for sustaining self-renewal of ground-state pluripotent stem cells.Nat Commun. 2021 May 24;12(1):3034. doi: 10.1038/s41467-021-23308-4. Nat Commun. 2021. PMID: 34031396 Free PMC article.
-
Generalized nuclear localization of retroelement transcripts.Mob DNA. 2022 Dec 2;13(1):30. doi: 10.1186/s13100-022-00287-x. Mob DNA. 2022. PMID: 36461093 Free PMC article.
-
2-Cell-like Cells: An Avenue for Improving SCNT Efficiency.Biomolecules. 2022 Nov 1;12(11):1611. doi: 10.3390/biom12111611. Biomolecules. 2022. PMID: 36358959 Free PMC article. Review.
-
Kick-starting the zygotic genome: licensors, specifiers, and beyond.EMBO Rep. 2024 Oct;25(10):4113-4130. doi: 10.1038/s44319-024-00223-5. Epub 2024 Aug 19. EMBO Rep. 2024. PMID: 39160344 Free PMC article. Review.
-
L1EM: a tool for accurate locus specific LINE-1 RNA quantification.Bioinformatics. 2020 Feb 15;36(4):1167-1173. doi: 10.1093/bioinformatics/btz724. Bioinformatics. 2020. PMID: 31584629 Free PMC article.
References
-
- Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev. 2006;73:279–87. - PubMed
-
- Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–12. - PubMed
-
- Bulut-Karslioglu A, Macrae TA, Oses-Prieto JA, Covarrubias S, Percharde M, Ku G, Diaz A, Mcmanus MT, Burlingame AL, Ramalho-Santos M. The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output. Cell Stem Cell. 2018;22:369–383. e8. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

