Episomal HBV persistence within transcribed host nuclear chromatin compartments involves HBx
- PMID: 29933745
- PMCID: PMC6015472
- DOI: 10.1186/s13072-018-0204-2
Episomal HBV persistence within transcribed host nuclear chromatin compartments involves HBx
Abstract
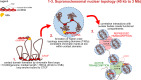
Background: In hepatocyte nuclei, hepatitis B virus (HBV) genomes occur episomally as covalently closed circular DNA (cccDNA). The HBV X protein (HBx) is required to initiate and maintain HBV replication. The functional nuclear localization of cccDNA and HBx remains unexplored.
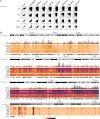
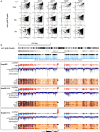
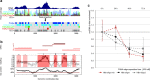
Results: To identify virus-host genome interactions and the underlying nuclear landscape for the first time, we combined circular chromosome conformation capture (4C) with RNA-seq and ChIP-seq. Moreover, we studied HBx-binding to HBV episomes. In HBV-positive HepaRG hepatocytes, we observed preferential association of HBV episomes and HBx with actively transcribed nuclear domains on the host genome correlating in size with constrained topological units of chromatin. Interestingly, HBx alone occupied transcribed chromatin domains. Silencing of native HBx caused reduced episomal HBV stability.
Conclusions: As part of the HBV episome, HBx might stabilize HBV episomal nuclear localization. Our observations may contribute to the understanding of long-term episomal stability and the facilitation of viral persistence. The exact mechanism by which HBx contributes to HBV nuclear persistence warrants further investigations.
Keywords: Chromatin fiber loops; Epigenome; Episome; HBxAg; Host–pathogen interaction; Oncogene; Supranucleosomal structure; TADs; Transcription factories; X-protein.
Figures

Similar articles
-
Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner.J Virol. 2019 Mar 5;93(6):e01840-18. doi: 10.1128/JVI.01840-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567987 Free PMC article.
-
Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection.J Hepatol. 2011 Nov;55(5):996-1003. doi: 10.1016/j.jhep.2011.02.015. Epub 2011 Mar 2. J Hepatol. 2011. PMID: 21376091
-
Inhibition of HBV Transcription From cccDNA With Nitazoxanide by Targeting the HBx-DDB1 Interaction.Cell Mol Gastroenterol Hepatol. 2019;7(2):297-312. doi: 10.1016/j.jcmgh.2018.10.010. Epub 2018 Oct 24. Cell Mol Gastroenterol Hepatol. 2019. PMID: 30704981 Free PMC article.
-
Identifying and Characterizing Interplay between Hepatitis B Virus X Protein and Smc5/6.Viruses. 2017 Apr 3;9(4):69. doi: 10.3390/v9040069. Viruses. 2017. PMID: 28368357 Free PMC article. Review.
-
Hepatitis B Virus X and Regulation of Viral Gene Expression.Cold Spring Harb Perspect Med. 2016 Jan 8;6(3):a021402. doi: 10.1101/cshperspect.a021402. Cold Spring Harb Perspect Med. 2016. PMID: 26747833 Free PMC article. Review.
Cited by
-
HBx represses WDR77 to enhance HBV replication by DDB1-mediated WDR77 degradation in the liver.Theranostics. 2021 Jul 25;11(17):8362-8378. doi: 10.7150/thno.57531. eCollection 2021. Theranostics. 2021. PMID: 34373747 Free PMC article.
-
Modification of Nuclear Compartments and the 3D Genome in the Course of a Viral Infection.Acta Naturae. 2020 Oct-Dec;12(4):34-46. doi: 10.32607/actanaturae.11041. Acta Naturae. 2020. PMID: 33456976 Free PMC article.
-
Topological implications of DNA tumor viral episomes.BMB Rep. 2022 Dec;55(12):587-594. doi: 10.5483/BMBRep.2022.55.12.154. BMB Rep. 2022. PMID: 36379513 Free PMC article. Review.
-
Imaging of Hepatitis B Virus Nucleic Acids: Current Advances and Challenges.Viruses. 2022 Mar 8;14(3):557. doi: 10.3390/v14030557. Viruses. 2022. PMID: 35336964 Free PMC article. Review.
-
Hepatitis B Virus HBx Protein Mediates the Degradation of Host Restriction Factors through the Cullin 4 DDB1 E3 Ubiquitin Ligase Complex.Cells. 2020 Mar 30;9(4):834. doi: 10.3390/cells9040834. Cells. 2020. PMID: 32235678 Free PMC article.
References
-
- WHO. WHO factsheet. 2015. http://wwwwhoint/mediacentre/factsheets/fs204/en/.
-
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

