Dual Roles for Ikaros in Regulation of Macrophage Chromatin State and Inflammatory Gene Expression
- PMID: 29898962
- PMCID: PMC6069956
- DOI: 10.4049/jimmunol.1800158
Dual Roles for Ikaros in Regulation of Macrophage Chromatin State and Inflammatory Gene Expression
Abstract
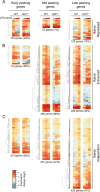
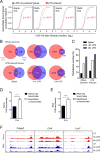
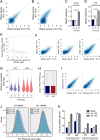
Macrophage activation by bacterial LPS leads to induction of a complex inflammatory gene program dependent on numerous transcription factor families. The transcription factor Ikaros has been shown to play a critical role in lymphoid cell development and differentiation; however, its function in myeloid cells and innate immune responses is less appreciated. Using comprehensive genomic analysis of Ikaros-dependent transcription, DNA binding, and chromatin accessibility, we describe unexpected dual repressor and activator functions for Ikaros in the LPS response of murine macrophages. Consistent with the described function of Ikaros as transcriptional repressor, Ikzf1-/- macrophages showed enhanced induction for select responses. In contrast, we observed a dramatic defect in expression of many delayed LPS response genes, and chromatin immunoprecipitation sequencing analyses support a key role for Ikaros in sustained NF-κB chromatin binding. Decreased Ikaros expression in Ikzf1+/- mice and human cells dampens these Ikaros-enhanced inflammatory responses, highlighting the importance of quantitative control of Ikaros protein level for its activator function. In the absence of Ikaros, a constitutively open chromatin state was coincident with dysregulation of LPS-induced chromatin remodeling, gene expression, and cytokine responses. Together, our data suggest a central role for Ikaros in coordinating the complex macrophage transcriptional program in response to pathogen challenge.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
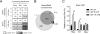


Similar articles
-
The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis.PLoS Genet. 2014 Dec 4;10(12):e1004827. doi: 10.1371/journal.pgen.1004827. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25474253 Free PMC article.
-
Ikaros antagonizes DNA binding by STAT5 in pre-B cells.PLoS One. 2020 Nov 12;15(11):e0242211. doi: 10.1371/journal.pone.0242211. eCollection 2020. PLoS One. 2020. PMID: 33180866 Free PMC article.
-
Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development.J Biol Chem. 2013 Dec 6;288(49):35170-9. doi: 10.1074/jbc.M113.481440. Epub 2013 Oct 21. J Biol Chem. 2013. PMID: 24145030 Free PMC article.
-
Ikaros and tumor suppression in acute lymphoblastic leukemia.Crit Rev Oncog. 2011;16(1-2):3-12. doi: 10.1615/critrevoncog.v16.i1-2.20. Crit Rev Oncog. 2011. PMID: 22150303 Free PMC article. Review.
-
Multiple functions of Ikaros in hematological malignancies, solid tumor and autoimmune diseases.Gene. 2019 Feb 5;684:47-52. doi: 10.1016/j.gene.2018.10.045. Epub 2018 Oct 22. Gene. 2019. PMID: 30352248 Review.
Cited by
-
A single ChIP-seq dataset is sufficient for comprehensive analysis of motifs co-occurrence with MCOT package.Nucleic Acids Res. 2019 Dec 2;47(21):e139. doi: 10.1093/nar/gkz800. Nucleic Acids Res. 2019. PMID: 31750523 Free PMC article.
-
CCL2 mediated IKZF1 expression promotes M2 polarization of glioma-associated macrophages through CD84-SHP2 pathway.Oncogene. 2024 Aug;43(36):2737-2749. doi: 10.1038/s41388-024-03118-w. Epub 2024 Aug 7. Oncogene. 2024. PMID: 39112517
-
Attenuated Negative Feedback in Monocyte-Derived Macrophages From Persons Living With HIV: A Role for IKAROS.Front Immunol. 2021 Nov 30;12:785905. doi: 10.3389/fimmu.2021.785905. eCollection 2021. Front Immunol. 2021. PMID: 34917094 Free PMC article.
-
Changes in H3K27ac at Gene Regulatory Regions in Porcine Alveolar Macrophages Following LPS or PolyIC Exposure.Front Genet. 2020 Aug 20;11:817. doi: 10.3389/fgene.2020.00817. eCollection 2020. Front Genet. 2020. PMID: 32973863 Free PMC article.
-
Nano-Antagonist Alleviates Inflammation and Allows for MRI of Atherosclerosis.Nanotheranostics. 2019 Nov 1;3(4):342-355. doi: 10.7150/ntno.37391. eCollection 2019. Nanotheranostics. 2019. PMID: 31723548 Free PMC article.
References
-
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. - PubMed
-
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. - PubMed
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

