The Nutrient-Sensing Hexosamine Biosynthetic Pathway as the Hub of Cancer Metabolic Rewiring
- PMID: 29865240
- PMCID: PMC6025041
- DOI: 10.3390/cells7060053
The Nutrient-Sensing Hexosamine Biosynthetic Pathway as the Hub of Cancer Metabolic Rewiring
Abstract
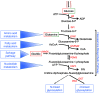
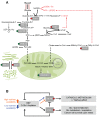
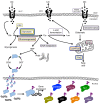
Alterations in glucose and glutamine utilizing pathways and in fatty acid metabolism are currently considered the most significant and prevalent metabolic changes observed in almost all types of tumors. Glucose, glutamine and fatty acids are the substrates for the hexosamine biosynthetic pathway (HBP). This metabolic pathway generates the "sensing molecule" UDP-N-Acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is the substrate for the enzymes involved in protein N- and O-glycosylation, two important post-translational modifications (PTMs) identified in several proteins localized in the extracellular space, on the cell membrane and in the cytoplasm, nucleus and mitochondria. Since protein glycosylation controls several key aspects of cell physiology, aberrant protein glycosylation has been associated with different human diseases, including cancer. Here we review recent evidence indicating the tight association between the HBP flux and cell metabolism, with particular emphasis on the post-transcriptional and transcriptional mechanisms regulated by the HBP that may cause the metabolic rewiring observed in cancer. We describe the implications of both protein O- and N-glycosylation in cancer cell metabolism and bioenergetics; focusing our attention on the effect of these PTMs on nutrient transport and on the transcriptional regulation and function of cancer-specific metabolic pathways.
Keywords: UDP-GlcNAc; bioenergetics; cancer; hexosamine biosynthesis pathway; metabolism; protein glycosylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Epithelial Mesenchymal Transition Induces Aberrant Glycosylation through Hexosamine Biosynthetic Pathway Activation.J Biol Chem. 2016 Jun 17;291(25):12917-29. doi: 10.1074/jbc.M116.729236. Epub 2016 Apr 18. J Biol Chem. 2016. PMID: 27129262 Free PMC article.
-
First characterization of glucose flux through the hexosamine biosynthesis pathway (HBP) in ex vivo mouse heart.J Biol Chem. 2020 Feb 14;295(7):2018-2033. doi: 10.1074/jbc.RA119.010565. Epub 2020 Jan 8. J Biol Chem. 2020. PMID: 31915250 Free PMC article.
-
Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer.BMC Biol. 2019 Jul 4;17(1):52. doi: 10.1186/s12915-019-0671-3. BMC Biol. 2019. PMID: 31272438 Free PMC article. Review.
-
The Hexosamine Biosynthesis Pathway: Regulation and Function.Genes (Basel). 2023 Apr 18;14(4):933. doi: 10.3390/genes14040933. Genes (Basel). 2023. PMID: 37107691 Free PMC article. Review.
-
Glutamine deprivation triggers NAGK-dependent hexosamine salvage.Elife. 2021 Nov 30;10:e62644. doi: 10.7554/eLife.62644. Elife. 2021. PMID: 34844667 Free PMC article.
Cited by
-
Glycosylation in health and disease.Nat Rev Nephrol. 2019 Jun;15(6):346-366. doi: 10.1038/s41581-019-0129-4. Nat Rev Nephrol. 2019. PMID: 30858582 Free PMC article. Review.
-
Apoptosis of cancer cells is triggered by selective crosslinking and inhibition of receptor tyrosine kinases.Commun Biol. 2019 Jun 21;2:231. doi: 10.1038/s42003-019-0484-5. eCollection 2019. Commun Biol. 2019. PMID: 31263775 Free PMC article.
-
O-GlcNAcylation regulation of cellular signaling in cancer.Cell Signal. 2022 Feb;90:110201. doi: 10.1016/j.cellsig.2021.110201. Epub 2021 Nov 17. Cell Signal. 2022. PMID: 34800629 Free PMC article. Review.
-
The Warburg effect is necessary to promote glycosylation in the blastema during zebrafish tail regeneration.NPJ Regen Med. 2021 Sep 13;6(1):55. doi: 10.1038/s41536-021-00163-x. NPJ Regen Med. 2021. PMID: 34518542 Free PMC article.
-
Hexosamine biosynthesis and related pathways, protein N-glycosylation and O-GlcNAcylation: their interconnection and role in plants.Front Plant Sci. 2024 Mar 6;15:1349064. doi: 10.3389/fpls.2024.1349064. eCollection 2024. Front Plant Sci. 2024. PMID: 38510444 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

