Lipid metabolism reprogramming and its potential targets in cancer
- PMID: 29784041
- PMCID: PMC5993136
- DOI: 10.1186/s40880-018-0301-4
Lipid metabolism reprogramming and its potential targets in cancer
Abstract
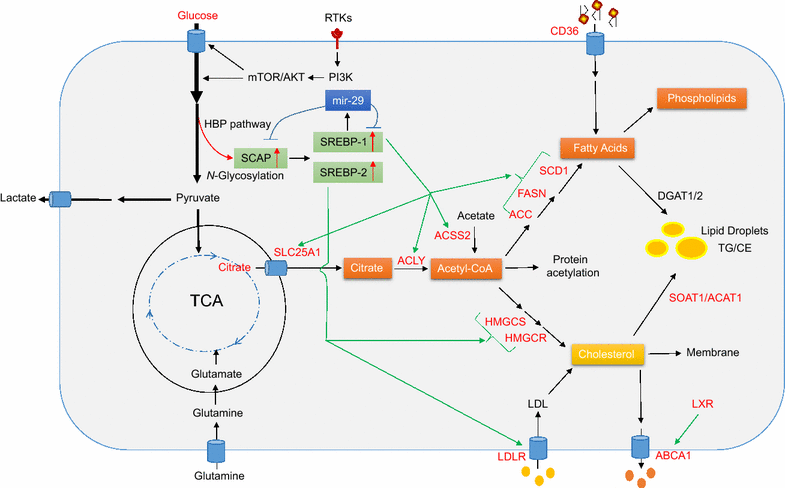
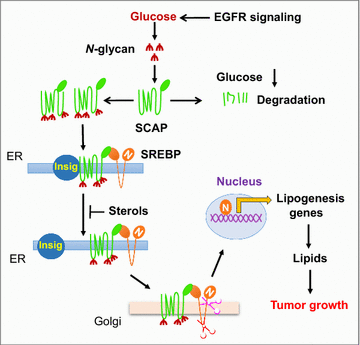
Reprogramming of lipid metabolism is a newly recognized hallmark of malignancy. Increased lipid uptake, storage and lipogenesis occur in a variety of cancers and contribute to rapid tumor growth. Lipids constitute the basic structure of membranes and also function as signaling molecules and energy sources. Sterol regulatory element-binding proteins (SREBPs), a family of membrane-bound transcription factors in the endoplasmic reticulum, play a central role in the regulation of lipid metabolism. Recent studies have revealed that SREBPs are highly up-regulated in various cancers and promote tumor growth. SREBP cleavage-activating protein is a key transporter in the trafficking and activation of SREBPs as well as a critical glucose sensor, thus linking glucose metabolism and de novo lipid synthesis. Targeting altered lipid metabolic pathways has become a promising anti-cancer strategy. This review summarizes recent progress in our understanding of lipid metabolism regulation in malignancy, and highlights potential molecular targets and their inhibitors for cancer treatment.
Keywords: Cancer; Cholesterol; Fatty acids; Lipid droplets; Lipid metabolism; SCAP; SREBPs.
Figures
Similar articles
-
SCAP/SREBPs are Central Players in Lipid Metabolism and Novel Metabolic Targets in Cancer Therapy.Curr Top Med Chem. 2018;18(6):484-493. doi: 10.2174/1568026618666180523104541. Curr Top Med Chem. 2018. PMID: 29788888 Free PMC article. Review.
-
Dipyridamole Inhibits Lipogenic Gene Expression by Retaining SCAP-SREBP in the Endoplasmic Reticulum.Cell Chem Biol. 2021 Feb 18;28(2):169-179.e7. doi: 10.1016/j.chembiol.2020.10.003. Epub 2020 Oct 22. Cell Chem Biol. 2021. PMID: 33096051 Free PMC article.
-
Heat Shock Protein 90 Modulates Lipid Homeostasis by Regulating the Stability and Function of Sterol Regulatory Element-binding Protein (SREBP) and SREBP Cleavage-activating Protein.J Biol Chem. 2017 Feb 17;292(7):3016-3028. doi: 10.1074/jbc.M116.767277. Epub 2016 Dec 21. J Biol Chem. 2017. PMID: 28003358 Free PMC article.
-
COPI-mediated retrieval of SCAP is crucial for regulating lipogenesis under basal and sterol-deficient conditions.J Cell Sci. 2015 Aug 1;128(15):2805-15. doi: 10.1242/jcs.164137. Epub 2015 Jun 19. J Cell Sci. 2015. PMID: 26092941
-
Sterol regulatory element-binding protein family as global regulators of lipid synthetic genes in energy metabolism.Vitam Horm. 2002;65:167-94. doi: 10.1016/s0083-6729(02)65064-2. Vitam Horm. 2002. PMID: 12481547 Review.
Cited by
-
Targeting SREBP-2-Regulated Mevalonate Metabolism for Cancer Therapy.Front Oncol. 2020 Aug 21;10:1510. doi: 10.3389/fonc.2020.01510. eCollection 2020. Front Oncol. 2020. PMID: 32974183 Free PMC article. Review.
-
SREBP1/FASN/cholesterol axis facilitates radioresistance in colorectal cancer.FEBS Open Bio. 2021 May;11(5):1343-1352. doi: 10.1002/2211-5463.13137. Epub 2021 May 1. FEBS Open Bio. 2021. PMID: 33665967 Free PMC article.
-
The role of lipid metabolism in tumor immune microenvironment and potential therapeutic strategies.Front Oncol. 2022 Sep 12;12:984560. doi: 10.3389/fonc.2022.984560. eCollection 2022. Front Oncol. 2022. PMID: 36172157 Free PMC article. Review.
-
Tissue metabolomics identified new biomarkers for the diagnosis and prognosis prediction of pancreatic cancer.Front Oncol. 2022 Sep 2;12:991051. doi: 10.3389/fonc.2022.991051. eCollection 2022. Front Oncol. 2022. PMID: 36119530 Free PMC article.
-
Shaping of Dendritic Cell Function by the Metabolic Micro-Environment.Front Endocrinol (Lausanne). 2020 Aug 28;11:555. doi: 10.3389/fendo.2020.00555. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33013685 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

