Emerging Evidence of Chromosome Folding by Loop Extrusion
- PMID: 29728444
- PMCID: PMC6512960
- DOI: 10.1101/sqb.2017.82.034710
Emerging Evidence of Chromosome Folding by Loop Extrusion
Abstract
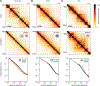
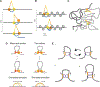
Chromosome organization poses a remarkable physical problem with many biological consequences: How can molecular interactions between proteins at the nanometer scale organize micron-long chromatinized DNA molecules, insulating or facilitating interactions between specific genomic elements? The mechanism of active loop extrusion holds great promise for explaining interphase and mitotic chromosome folding, yet remains difficult to assay directly. We discuss predictions from our polymer models of loop extrusion with barrier elements and review recent experimental studies that provide strong support for loop extrusion, focusing on perturbations to CTCF and cohesin assayed via Hi-C in interphase. Finally, we discuss a likely molecular mechanism of loop extrusion by structural maintenance of chromosomes complexes.
© 2017 Fudenberg et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
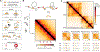

Similar articles
-
Chromosome organization by one-sided and two-sided loop extrusion.Elife. 2020 Apr 6;9:e53558. doi: 10.7554/eLife.53558. Elife. 2020. PMID: 32250245 Free PMC article.
-
Hit the brakes - a new perspective on the loop extrusion mechanism of cohesin and other SMC complexes.J Cell Sci. 2021 Jan 8;134(1):jcs247577. doi: 10.1242/jcs.247577. J Cell Sci. 2021. PMID: 33419949 Review.
-
Formation of Chromosomal Domains by Loop Extrusion.Cell Rep. 2016 May 31;15(9):2038-49. doi: 10.1016/j.celrep.2016.04.085. Epub 2016 May 19. Cell Rep. 2016. PMID: 27210764 Free PMC article.
-
Genome organization via loop extrusion, insights from polymer physics models.Brief Funct Genomics. 2020 Mar 23;19(2):119-127. doi: 10.1093/bfgp/elz023. Brief Funct Genomics. 2020. PMID: 31711163
-
Two major mechanisms of chromosome organization.Curr Opin Cell Biol. 2019 Jun;58:142-152. doi: 10.1016/j.ceb.2019.05.001. Epub 2019 Jun 20. Curr Opin Cell Biol. 2019. PMID: 31228682 Free PMC article. Review.
Cited by
-
Differential contribution of steady-state RNA and active transcription in chromatin organization.EMBO Rep. 2019 Oct 4;20(10):e48068. doi: 10.15252/embr.201948068. Epub 2019 Aug 26. EMBO Rep. 2019. PMID: 31448565 Free PMC article.
-
The histone H4 lysine 20 demethylase DPY-21 regulates the dynamics of condensin DC binding.J Cell Sci. 2022 Jan 15;135(2):jcs258818. doi: 10.1242/jcs.258818. Epub 2022 Jan 26. J Cell Sci. 2022. PMID: 34918745 Free PMC article.
-
Moderation of Structural DNA Properties by Coupled Dinucleotide Contents in Eukaryotes.Genes (Basel). 2023 Mar 20;14(3):755. doi: 10.3390/genes14030755. Genes (Basel). 2023. PMID: 36981025 Free PMC article.
-
Smc3 dosage regulates B cell transit through germinal centers and restricts their malignant transformation.Nat Immunol. 2021 Feb;22(2):240-253. doi: 10.1038/s41590-020-00827-8. Epub 2021 Jan 11. Nat Immunol. 2021. PMID: 33432228 Free PMC article.
-
Quantitative prediction of enhancer-promoter interactions.Genome Res. 2020 Jan;30(1):72-84. doi: 10.1101/gr.249367.119. Epub 2019 Dec 2. Genome Res. 2020. PMID: 31804952 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
