Mechanical unfolding reveals stable 3-helix intermediates in talin and α-catenin
- PMID: 29698481
- PMCID: PMC5940241
- DOI: 10.1371/journal.pcbi.1006126
Mechanical unfolding reveals stable 3-helix intermediates in talin and α-catenin
Abstract
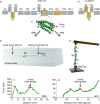
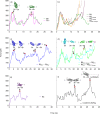
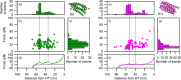
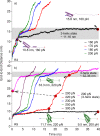
Mechanical stability is a key feature in the regulation of structural scaffolding proteins and their functions. Despite the abundance of α-helical structures among the human proteome and their undisputed importance in health and disease, the fundamental principles of their behavior under mechanical load are poorly understood. Talin and α-catenin are two key molecules in focal adhesions and adherens junctions, respectively. In this study, we used a combination of atomistic steered molecular dynamics (SMD) simulations, polyprotein engineering, and single-molecule atomic force microscopy (smAFM) to investigate unfolding of these proteins. SMD simulations revealed that talin rod α-helix bundles as well as α-catenin α-helix domains unfold through stable 3-helix intermediates. While the 5-helix bundles were found to be mechanically stable, a second stable conformation corresponding to the 3-helix state was revealed. Mechanically weaker 4-helix bundles easily unfolded into a stable 3-helix conformation. The results of smAFM experiments were in agreement with the findings of the computational simulations. The disulfide clamp mutants, designed to protect the stable state, support the 3-helix intermediate model in both experimental and computational setups. As a result, multiple discrete unfolding intermediate states in the talin and α-catenin unfolding pathway were discovered. Better understanding of the mechanical unfolding mechanism of α-helix proteins is a key step towards comprehensive models describing the mechanoregulation of proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Molecular dynamics simulations reveal how vinculin refolds partially unfolded talin rod helices to stabilize them against mechanical force.PLoS Comput Biol. 2024 Aug 7;20(8):e1012341. doi: 10.1371/journal.pcbi.1012341. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39110765 Free PMC article.
-
All Subdomains of the Talin Rod Are Mechanically Vulnerable and May Contribute To Cellular Mechanosensing.ACS Nano. 2016 Jul 26;10(7):6648-58. doi: 10.1021/acsnano.6b01658. Epub 2016 Jul 11. ACS Nano. 2016. PMID: 27380548 Free PMC article.
-
Nano-mechanical characterization of tension-sensitive helix bundles in talin rod.Biochem Biophys Res Commun. 2017 Mar 4;484(2):372-377. doi: 10.1016/j.bbrc.2017.01.127. Epub 2017 Jan 25. Biochem Biophys Res Commun. 2017. PMID: 28131835
-
Uncovering mechanosensing mechanisms at the single protein level using magnetic tweezers.Methods. 2016 Feb 1;94:13-8. doi: 10.1016/j.ymeth.2015.08.020. Epub 2015 Aug 28. Methods. 2016. PMID: 26318089 Review.
-
Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering.Prog Biophys Mol Biol. 2000;74(1-2):63-91. doi: 10.1016/s0079-6107(00)00017-1. Prog Biophys Mol Biol. 2000. PMID: 11106807 Review.
Cited by
-
Cancer associated talin point mutations disorganise cell adhesion and migration.Sci Rep. 2021 Jan 11;11(1):347. doi: 10.1038/s41598-020-77911-4. Sci Rep. 2021. PMID: 33431906 Free PMC article.
-
Molecular dynamics simulations reveal how vinculin refolds partially unfolded talin rod helices to stabilize them against mechanical force.PLoS Comput Biol. 2024 Aug 7;20(8):e1012341. doi: 10.1371/journal.pcbi.1012341. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39110765 Free PMC article.
-
Talin dissociates from RIAM and associates to vinculin sequentially in response to the actomyosin force.Nat Commun. 2020 Jun 19;11(1):3116. doi: 10.1038/s41467-020-16922-1. Nat Commun. 2020. PMID: 32561773 Free PMC article.
-
Cell-cell adhesion interface: orthogonal and parallel forces from contraction, protrusion, and retraction.F1000Res. 2018 Sep 25;7:F1000 Faculty Rev-1544. doi: 10.12688/f1000research.15860.1. eCollection 2018. F1000Res. 2018. PMID: 30345009 Free PMC article. Review.
-
The mechanical cell - the role of force dependencies in synchronising protein interaction networks.J Cell Sci. 2022 Nov 15;135(22):jcs259769. doi: 10.1242/jcs.259769. Epub 2022 Nov 18. J Cell Sci. 2022. PMID: 36398718 Free PMC article. Review.
References
-
- Seifert C, Grater F. Protein mechanics: how force regulates molecular function. BiochimBiophysActa. 2013;1830: 4762–4768. doi: 10.1016/j.bbagen.2013.06.005 - DOI - PubMed
-
- Hu X, Margadant FM, Yao M, Sheetz MP. Molecular stretching modulates mechanosensing pathways. Protein Sci. 2017;26: 1337–1351. doi: 10.1002/pro.3188 - DOI - PMC - PubMed
-
- Ipsaro JJ, Harper SL, Messick TE, Marmorstein R, Mondragon A, Speicher DW. Crystal structure and functional interpretation of the erythrocyte spectrintetramerization domain complex. Blood. 2010;115: 4843–4852. doi: 10.1182/blood-2010-01-261396 - DOI - PMC - PubMed
-
- Muthu M, Richardson KA, Sutherland-Smith AJ. The crystal structures of dystrophin and utrophinspectrin repeats: implications for domain boundaries. PLoS One. 2012;7: e40066 doi: 10.1371/journal.pone.0040066 - DOI - PMC - PubMed
-
- Choi H-J, Weis WI. Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakinplakin domain. J Mol Biol. 2011;409: 800–812. doi: 10.1016/j.jmb.2011.04.046 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

