RNA N6-methyladenosine modification in cancers: current status and perspectives
- PMID: 29686311
- PMCID: PMC5951805
- DOI: 10.1038/s41422-018-0034-6
RNA N6-methyladenosine modification in cancers: current status and perspectives
Abstract
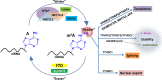
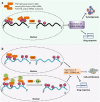
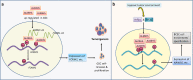
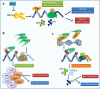
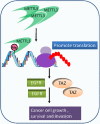
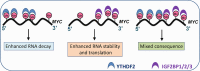
N6-methyladenosine (m6A), the most abundant internal modification in eukaryotic messenger RNAs (mRNAs), has been shown to play critical roles in various normal bioprocesses such as tissue development, stem cell self-renewal and differentiation, heat shock or DNA damage response, and maternal-to-zygotic transition. The m6A modification is deposited by the m6A methyltransferase complex (MTC; i.e., writer) composed of METTL3, METTL14 and WTAP, and probably also VIRMA and RBM15, and can be removed by m6A demethylases (i.e., erasers) such as FTO and ALKBH5. The fates of m6A-modified mRNAs rely on the functions of distinct proteins that recognize them (i.e., readers), which may affect the stability, splicing, and/or translation of target mRNAs. Given the functional importance of the m6A modification machinery in normal bioprocesses, it is not surprising that evidence is emerging that dysregulation of m6A modification and the associated proteins also contributes to the initiation, progression, and drug response of cancers. In this review, we focus on recent advances in the study of biological functions and the underlying molecular mechanisms of dysregulated m6A modification and the associated machinery in the pathogenesis and drug response of various types of cancers. In addition, we also discuss possible therapeutic interventions against the dysregulated m6A machinery to treat cancers.
Conflict of interest statement
We have a patent filed based on our R-2HG/FTO work (to J.C. and R.S.). The remaining authors declare no competing interests.
Figures

Similar articles
-
RNA N 6-Methyladenosine Modification in Normal and Malignant Hematopoiesis.Adv Exp Med Biol. 2019;1143:75-93. doi: 10.1007/978-981-13-7342-8_4. Adv Exp Med Biol. 2019. PMID: 31338816 Review.
-
RNA N6-methyladenosine modification in solid tumors: new therapeutic frontiers.Cancer Gene Ther. 2020 Sep;27(9):625-633. doi: 10.1038/s41417-020-0160-4. Epub 2020 Jan 20. Cancer Gene Ther. 2020. PMID: 31956264 Free PMC article. Review.
-
Function and evolution of RNA N6-methyladenosine modification.Int J Biol Sci. 2020 Apr 15;16(11):1929-1940. doi: 10.7150/ijbs.45231. eCollection 2020. Int J Biol Sci. 2020. PMID: 32398960 Free PMC article. Review.
-
The Role of Dynamic m6 A RNA Methylation in Photobiology.Photochem Photobiol. 2019 Jan;95(1):95-104. doi: 10.1111/php.12930. Epub 2018 May 24. Photochem Photobiol. 2019. PMID: 29729018 Free PMC article. Review.
-
The role of m6A RNA methylation in human cancer.Mol Cancer. 2019 May 29;18(1):103. doi: 10.1186/s12943-019-1033-z. Mol Cancer. 2019. PMID: 31142332 Free PMC article. Review.
Cited by
-
Wilms tumor 1 associated protein promotes epithelial mesenchymal transition of gastric cancer cells by accelerating TGF-β and enhances chemoradiotherapy resistance.J Cancer Res Clin Oncol. 2023 Jul;149(7):3977-3988. doi: 10.1007/s00432-022-04320-7. Epub 2022 Aug 28. J Cancer Res Clin Oncol. 2023. PMID: 36030434
-
RBM15 Protects From Myocardial Infarction by Stabilizing NAE1.JACC Basic Transl Sci. 2024 Apr 10;9(5):631-648. doi: 10.1016/j.jacbts.2024.01.017. eCollection 2024 May. JACC Basic Transl Sci. 2024. PMID: 38984049 Free PMC article.
-
m6A Methylation-Mediated Stabilization of LINC01106 Suppresses Bladder Cancer Progression by Regulating the miR-3148/DAB1 Axis.Biomedicines. 2024 Jan 5;12(1):114. doi: 10.3390/biomedicines12010114. Biomedicines. 2024. PMID: 38255219 Free PMC article.
-
Roles of METTL3 in cancer: mechanisms and therapeutic targeting.J Hematol Oncol. 2020 Aug 27;13(1):117. doi: 10.1186/s13045-020-00951-w. J Hematol Oncol. 2020. PMID: 32854717 Free PMC article. Review.
-
Modification of m6A mediates tissue immune microenvironment in calcific aortic valve disease.Ann Transl Med. 2022 Sep;10(17):931. doi: 10.21037/atm-22-3627. Ann Transl Med. 2022. PMID: 36172101 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

