Acetyl-CoA promotes glioblastoma cell adhesion and migration through Ca2+-NFAT signaling
- PMID: 29674394
- PMCID: PMC5959234
- DOI: 10.1101/gad.311027.117
Acetyl-CoA promotes glioblastoma cell adhesion and migration through Ca2+-NFAT signaling
Abstract
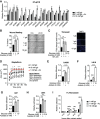
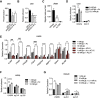
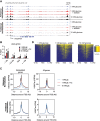
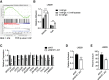
The metabolite acetyl-coenzyme A (acetyl-CoA) is the required acetyl donor for lysine acetylation and thereby links metabolism, signaling, and epigenetics. Nutrient availability alters acetyl-CoA levels in cancer cells, correlating with changes in global histone acetylation and gene expression. However, the specific molecular mechanisms through which acetyl-CoA production impacts gene expression and its functional roles in promoting malignant phenotypes are poorly understood. Here, using histone H3 Lys27 acetylation (H3K27ac) ChIP-seq (chromatin immunoprecipitation [ChIP] coupled with next-generation sequencing) with normalization to an exogenous reference genome (ChIP-Rx), we found that changes in acetyl-CoA abundance trigger site-specific regulation of H3K27ac, correlating with gene expression as opposed to uniformly modulating this mark at all genes. Genes involved in integrin signaling and cell adhesion were identified as acetyl-CoA-responsive in glioblastoma cells, and we demonstrate that ATP citrate lyase (ACLY)-dependent acetyl-CoA production promotes cell migration and adhesion to the extracellular matrix. Mechanistically, the transcription factor NFAT1 (nuclear factor of activated T cells 1) was found to mediate acetyl-CoA-dependent gene regulation and cell adhesion. This occurs through modulation of Ca2+ signals, triggering NFAT1 nuclear translocation when acetyl-CoA is abundant. The findings of this study thus establish that acetyl-CoA impacts H3K27ac at specific loci, correlating with gene expression, and that expression of cell adhesion genes are driven by acetyl-CoA in part through activation of Ca2+-NFAT signaling.
Keywords: NFAT1; acetyl-CoA; calcium; glioblastoma; histone acetylation; metabolism.
© 2018 Lee et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Comment in
-
Acetyl-CoA-directed gene transcription in cancer cells.Genes Dev. 2018 Apr 1;32(7-8):463-465. doi: 10.1101/gad.315168.118. Genes Dev. 2018. PMID: 29692354 Free PMC article.
Similar articles
-
Mammalian SIRT6 Represses Invasive Cancer Cell Phenotypes through ATP Citrate Lyase (ACLY)-Dependent Histone Acetylation.Genes (Basel). 2021 Sep 21;12(9):1460. doi: 10.3390/genes12091460. Genes (Basel). 2021. PMID: 34573442 Free PMC article.
-
Modulation of matrix metabolism by ATP-citrate lyase in articular chondrocytes.J Biol Chem. 2018 Aug 3;293(31):12259-12270. doi: 10.1074/jbc.RA118.002261. Epub 2018 Jun 21. J Biol Chem. 2018. PMID: 29929979 Free PMC article.
-
Acetyl-CoA-directed gene transcription in cancer cells.Genes Dev. 2018 Apr 1;32(7-8):463-465. doi: 10.1101/gad.315168.118. Genes Dev. 2018. PMID: 29692354 Free PMC article.
-
Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination.Mol Cell. 2017 Jul 20;67(2):252-265.e6. doi: 10.1016/j.molcel.2017.06.008. Epub 2017 Jul 6. Mol Cell. 2017. PMID: 28689661 Free PMC article.
-
Exploring the Role of ATP-Citrate Lyase in the Immune System.Front Immunol. 2021 Feb 18;12:632526. doi: 10.3389/fimmu.2021.632526. eCollection 2021. Front Immunol. 2021. PMID: 33679780 Free PMC article. Review.
Cited by
-
Glioma glycolipid metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential treatment target.Clin Transl Med. 2021 May;11(5):e411. doi: 10.1002/ctm2.411. Clin Transl Med. 2021. PMID: 34047477 Free PMC article.
-
Biomaterial-Based Metabolic Regulation in Regenerative Engineering.Adv Sci (Weinh). 2019 Jul 28;6(19):1900819. doi: 10.1002/advs.201900819. eCollection 2019 Oct 2. Adv Sci (Weinh). 2019. PMID: 31592416 Free PMC article. Review.
-
Mitochondrial TCA cycle metabolites control physiology and disease.Nat Commun. 2020 Jan 3;11(1):102. doi: 10.1038/s41467-019-13668-3. Nat Commun. 2020. PMID: 31900386 Free PMC article. Review.
-
Increased proton-sensing receptor GPR4 signalling promotes colorectal cancer progression by activating the hippo pathway.EBioMedicine. 2019 Oct;48:264-276. doi: 10.1016/j.ebiom.2019.09.016. Epub 2019 Sep 14. EBioMedicine. 2019. PMID: 31530502 Free PMC article.
-
A Systems Chemoproteomic Analysis of Acyl-CoA/Protein Interaction Networks.Cell Chem Biol. 2020 Mar 19;27(3):322-333.e5. doi: 10.1016/j.chembiol.2019.11.011. Epub 2019 Dec 10. Cell Chem Biol. 2020. PMID: 31836350 Free PMC article.
References
-
- Badran BM, Wolinsky SM, Burny A, Willard-Gallo KE. 2002. Identification of three NFAT binding motifs in the 5′-upstream region of the human CD3γ gene that differentially bind NFATc1, NFATc2, and NF-κB p50. J Biol Chem 277: 47136–47148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
