Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis
- PMID: 29657300
- PMCID: PMC5890390
- DOI: 10.3390/ncrna4010003
Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis
Abstract
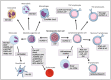
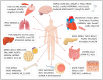
A significant proportion of mammalian genomes corresponds to genes that transcribe long non-coding RNAs (lncRNAs). Throughout the last decade, the number of studies concerning the roles played by lncRNAs in different biological processes has increased considerably. This intense interest in lncRNAs has produced a major shift in our understanding of gene and genome regulation and structure. It became apparent that lncRNAs regulate gene expression through several mechanisms. These RNAs function as transcriptional or post-transcriptional regulators through binding to histone-modifying complexes, to DNA, to transcription factors and other DNA binding proteins, to RNA polymerase II, to mRNA, or through the modulation of microRNA or enzyme function. Often, the lncRNA transcription itself rather than the lncRNA product appears to be regulatory. In this review, we highlight studies identifying lncRNAs in the homeostasis of various cell and tissue types or demonstrating their effects in the expression of protein-coding or other non-coding RNA genes.
Keywords: gene expression; gene regulation; homeostasis; long non-coding RNA; physiological regulatory mechanisms; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring.Crit Rev Clin Lab Sci. 2014 Dec;51(6):344-57. doi: 10.3109/10408363.2014.944299. Epub 2014 Aug 15. Crit Rev Clin Lab Sci. 2014. PMID: 25123609 Review.
-
Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice.BMC Genomics. 2018 Nov 21;19(1):834. doi: 10.1186/s12864-018-5235-3. BMC Genomics. 2018. PMID: 30463508 Free PMC article.
-
Long Non-Coding RNAs in Metabolic Organs and Energy Homeostasis.Int J Mol Sci. 2017 Nov 30;18(12):2578. doi: 10.3390/ijms18122578. Int J Mol Sci. 2017. PMID: 29189723 Free PMC article. Review.
-
How do lncRNAs regulate transcription?Sci Adv. 2017 Sep 27;3(9):eaao2110. doi: 10.1126/sciadv.aao2110. eCollection 2017 Sep. Sci Adv. 2017. PMID: 28959731 Free PMC article. Review.
-
Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress.BMC Plant Biol. 2018 Jan 25;18(1):23. doi: 10.1186/s12870-018-1238-0. BMC Plant Biol. 2018. PMID: 29370759 Free PMC article.
Cited by
-
Spontaneous evolution of human skin fibroblasts into wound-healing keratinocyte-like cells.Theranostics. 2019 Jul 9;9(18):5200-5213. doi: 10.7150/thno.31526. eCollection 2019. Theranostics. 2019. PMID: 31410210 Free PMC article.
-
Non-Coding RNAs and Hepatitis C Virus-Induced Hepatocellular Carcinoma.Viruses. 2018 Oct 30;10(11):591. doi: 10.3390/v10110591. Viruses. 2018. PMID: 30380697 Free PMC article. Review.
-
LncRNA and mRNA expression profiles in brown adipose tissue of obesity-prone and obesity-resistant mice.iScience. 2022 Jul 20;25(8):104809. doi: 10.1016/j.isci.2022.104809. eCollection 2022 Aug 19. iScience. 2022. PMID: 35992072 Free PMC article.
-
Long noncoding RNA EMS connects c-Myc to cell cycle control and tumorigenesis.Proc Natl Acad Sci U S A. 2019 Jul 16;116(29):14620-14629. doi: 10.1073/pnas.1903432116. Epub 2019 Jul 1. Proc Natl Acad Sci U S A. 2019. PMID: 31262817 Free PMC article.
-
Phytochemicals as Modulators of Long Non-Coding RNAs and Inhibitors of Cancer-Related Carbonic Anhydrases.Int J Mol Sci. 2019 Jun 15;20(12):2939. doi: 10.3390/ijms20122939. Int J Mol Sci. 2019. PMID: 31208095 Free PMC article. Review.
References
-
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

