Limited immune surveillance in lymphoid tissue by cytolytic CD4+ T cells during health and HIV disease
- PMID: 29652923
- PMCID: PMC5919077
- DOI: 10.1371/journal.ppat.1006973
Limited immune surveillance in lymphoid tissue by cytolytic CD4+ T cells during health and HIV disease
Abstract
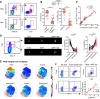
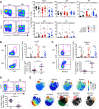
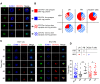
CD4+ T cells subsets have a wide range of important helper and regulatory functions in the immune system. Several studies have specifically suggested that circulating effector CD4+ T cells may play a direct role in control of HIV replication through cytolytic activity or autocrine β-chemokine production. However, it remains unclear whether effector CD4+ T cells expressing cytolytic molecules and β-chemokines are present within lymph nodes (LNs), a major site of HIV replication. Here, we report that expression of β-chemokines and cytolytic molecules are enriched within a CD4+ T cell population with high levels of the T-box transcription factors T-bet and eomesodermin (Eomes). This effector population is predominately found in peripheral blood and is limited in LNs regardless of HIV infection or treatment status. As a result, CD4+ T cells generally lack effector functions in LNs, including cytolytic capacity and IFNγ and β-chemokine expression, even in HIV elite controllers and during acute/early HIV infection. While we do find the presence of degranulating CD4+ T cells in LNs, these cells do not bear functional or transcriptional effector T cell properties and are inherently poor to form stable immunological synapses compared to their peripheral blood counterparts. We demonstrate that CD4+ T cell cytolytic function, phenotype, and programming in the peripheral blood is dissociated from those characteristics found in lymphoid tissues. Together, these data challenge our current models based on blood and suggest spatially and temporally dissociated mechanisms of viral control in lymphoid tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Decreased CXCR3+ CD8 T cells in advanced human immunodeficiency virus infection suggest that a homing defect contributes to cytotoxic T-lymphocyte dysfunction.J Virol. 2007 Aug;81(16):8439-50. doi: 10.1128/JVI.00199-07. Epub 2007 Jun 6. J Virol. 2007. PMID: 17553894 Free PMC article.
-
The role of secondary lymphatic tissue in immune deficiency of HIV infection.AIDS. 2008 Sep;22 Suppl 3:S13-8. doi: 10.1097/01.aids.0000327511.76126.b5. AIDS. 2008. PMID: 18845917
-
Elite control of HIV is associated with distinct functional and transcriptional signatures in lymphoid tissue CD8+ T cells.Sci Transl Med. 2019 Dec 18;11(523):eaax4077. doi: 10.1126/scitranslmed.aax4077. Sci Transl Med. 2019. PMID: 31852798 Free PMC article.
-
Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues.Annu Rev Immunol. 1999;17:625-56. doi: 10.1146/annurev.immunol.17.1.625. Annu Rev Immunol. 1999. PMID: 10358770 Review.
-
Role of lymphoid organs in the pathogenesis of human immunodeficiency virus (HIV) infection.Immunol Rev. 1994 Aug;140:105-30. doi: 10.1111/j.1600-065x.1994.tb00867.x. Immunol Rev. 1994. PMID: 7821924 Review.
Cited by
-
Regulation of CD4 T Cell Responses by the Transcription Factor Eomesodermin.Biomolecules. 2022 Oct 24;12(11):1549. doi: 10.3390/biom12111549. Biomolecules. 2022. PMID: 36358898 Free PMC article. Review.
-
The immune synapses reveal aberrant functions of CD8 T cells during chronic HIV infection.Nat Commun. 2022 Oct 28;13(1):6436. doi: 10.1038/s41467-022-34157-0. Nat Commun. 2022. PMID: 36307445 Free PMC article.
-
Risk and incidence of head and neck cancers in veterans living with HIV and matched HIV-negative veterans.Cancer. 2022 Sep 15;128(18):3310-3318. doi: 10.1002/cncr.34387. Epub 2022 Jul 22. Cancer. 2022. PMID: 35867552 Free PMC article.
-
Bone Mesenchymal Stem Cell-Derived Exosome-Enclosed miR-181a Induces CD4+CD25+FOXP3+ Regulatory T Cells via SIRT1/Acetylation-Mediated FOXP3 Stabilization.J Oncol. 2022 May 26;2022:8890434. doi: 10.1155/2022/8890434. eCollection 2022. J Oncol. 2022. PMID: 35664563 Free PMC article.
-
Granzyme B+ CD4 T cells accumulate in the colon during chronic HIV-1 infection.Gut Microbes. 2022 Jan-Dec;14(1):2045852. doi: 10.1080/19490976.2022.2045852. Gut Microbes. 2022. PMID: 35258402 Free PMC article.
References
-
- Cheroutre H, Husain MM. CD4 CTL: living up to the challenge. Semin Immunol. 2013;25(4):273–81. doi: 10.1016/j.smim.2013.10.022 ; PubMed Central PMCID: PMCPMC3886800. - DOI - PMC - PubMed
-
- Brown DM. Cytolytic CD4 cells: Direct mediators in infectious disease and malignancy. Cell Immunol. 2010;262(2):89–95. doi: 10.1016/j.cellimm.2010.02.008 ; PubMed Central PMCID: PMCPMC2874968. - DOI - PMC - PubMed
-
- Buggert M, Norstrom MM, Czarnecki C, Tupin E, Luo M, Gyllensten K, et al. Characterization of HIV-specific CD4+ T cell responses against peptides selected with broad population and pathogen coverage. PloS one. 2012;7(7):e39874 Epub 2012/07/14. doi: 10.1371/journal.pone.0039874 ; PubMed Central PMCID: PMC3390319. - DOI - PMC - PubMed
-
- Ferre AL, Hunt PW, McConnell DH, Morris MM, Garcia JC, Pollard RB, et al. HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses. J Virol. 2010;84(21):11020–9. doi: 10.1128/JVI.00980-10 ; PubMed Central PMCID: PMCPMC2953185. - DOI - PMC - PubMed
-
- Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, et al. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78(9):4463–77. Epub 2004/04/14. doi: 10.1128/JVI.78.9.4463-4477.2004 ; PubMed Central PMCID: PMC387674. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- UM1 AI126620/AI/NIAID NIH HHS/United States
- AI126620 /NH/NIH HHS/United States
- AI106481 /NH/NIH HHS/United States
- R01 AI108972/AI/NIAID NIH HHS/United States
- AI118694 /NH/NIH HHS/United States
- R01 AI118694/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- R56 AI106481/AI/NIAID NIH HHS/United States
- A127966 /NH/NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- R01 AI076066/AI/NIAID NIH HHS/United States
- AI076066 /NH/NIH HHS/United States
- U19 AI096109/AI/NIAID NIH HHS/United States
- P30 AI027763 /NH/NIH HHS/United States
- AI096109 /NH/NIH HHS/United States
- P30 AI036219/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

