Chimeric 3' flanking regions strongly enhance gene expression in plants
- PMID: 29637682
- PMCID: PMC6230951
- DOI: 10.1111/pbi.12931
Chimeric 3' flanking regions strongly enhance gene expression in plants
Abstract
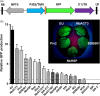
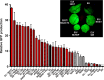
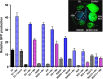
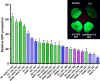
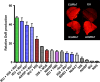
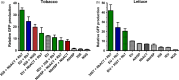
Plants represent a promising platform for the highly scalable production of recombinant proteins. Previously, we identified the tobacco extensin terminator lacking its intron as an element that reduced transcript read-through and improved recombinant protein production in a plant-based system. In this study, we systematically compared nonreplicating plant expression vectors containing over 20 commonly used or newly identified terminators from diverse sources. We found that eight gene terminators enhance reporter gene expression significantly more than the commonly used 35S and NOS terminators. The intronless extensin terminator provided a 13.6-fold increase compared with the NOS terminator. Combining terminators in tandem produced large synergistic effects, with many combinations providing a >25-fold increase in expression. Addition of the tobacco Rb7 or TM6 matrix attachment region (MAR) strongly enhanced protein production when added to most terminators, with the Rb7 MAR providing the greatest enhancement. Using deletion analysis, the full activity of the 1193 bp Rb7 MAR was found to require only a 463-bp region at its 3' end. Combined terminators and MAR together provided a >60-fold increase compared with the NOS terminator alone. These combinations were then placed in a replicating geminiviral vector, providing a total of >150-fold enhancement over the original NOS vector, corresponding to an estimated yield of 3-5 g recombinant protein per kg leaf fresh weight or around 50% of the leaf total soluble protein. These results demonstrate the importance of 3' flanking regions in optimizing gene expression and show great potential for 3' flanking regions to improve DNA-based recombinant protein production systems.
Keywords: 3’ UTR; biopharmaceutical; geminiviral vector; gene terminator; matrix attachment region; recombinant protein; transient expression.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

Similar articles
-
An intronless form of the tobacco extensin gene terminator strongly enhances transient gene expression in plant leaves.Plant Mol Biol. 2018 Mar;96(4-5):429-443. doi: 10.1007/s11103-018-0708-y. Epub 2018 Feb 10. Plant Mol Biol. 2018. PMID: 29429129
-
High-Level Production of a Recombinant Protein in Nicotiana benthamiana Leaves Through Transient Expression Using a Double Terminator.Int J Mol Sci. 2024 Oct 28;25(21):11573. doi: 10.3390/ijms252111573. Int J Mol Sci. 2024. PMID: 39519125 Free PMC article.
-
5' and 3' Untranslated Regions Strongly Enhance Performance of Geminiviral Replicons in Nicotiana benthamiana Leaves.Front Plant Sci. 2016 Feb 24;7:200. doi: 10.3389/fpls.2016.00200. eCollection 2016. Front Plant Sci. 2016. PMID: 26941764 Free PMC article.
-
A versatile vector system for multiple gene expression in plants.Trends Plant Sci. 2005 Aug;10(8):357-61. doi: 10.1016/j.tplants.2005.06.001. Trends Plant Sci. 2005. PMID: 15993643 Review.
-
Production of pharmaceutical proteins from transgenic animals.J Biotechnol. 1994 May 31;34(3):269-87. doi: 10.1016/0168-1656(94)90062-0. J Biotechnol. 1994. PMID: 7764960 Review.
Cited by
-
Identification of a Transferrable Terminator Element That Inhibits Small RNA Production and Improves Transgene Expression Levels.Front Plant Sci. 2022 May 16;13:877793. doi: 10.3389/fpls.2022.877793. eCollection 2022. Front Plant Sci. 2022. PMID: 35651775 Free PMC article.
-
Evolutionary conservation of nested MIR159 structural microRNA genes and their promoter characterization in Arabidopsis thaliana.Front Plant Sci. 2022 Jul 26;13:948751. doi: 10.3389/fpls.2022.948751. eCollection 2022. Front Plant Sci. 2022. PMID: 35958213 Free PMC article.
-
Plant Engineering to Enable Platforms for Sustainable Bioproduction of Terpenoids.Methods Mol Biol. 2024;2760:3-20. doi: 10.1007/978-1-0716-3658-9_1. Methods Mol Biol. 2024. PMID: 38468079 Review.
-
Insights obtained using different modules of the cotton uceA1.7 promoter.Planta. 2020 Jan 31;251(2):56. doi: 10.1007/s00425-020-03348-8. Planta. 2020. PMID: 32006110
-
Evaluation of a toxoid fusion protein vaccine produced in plants to protect poultry against necrotic enteritis.PeerJ. 2019 Mar 28;7:e6600. doi: 10.7717/peerj.6600. eCollection 2019. PeerJ. 2019. PMID: 30944775 Free PMC article.
References
-
- Allen, E. and Howell, M.D. (2010) miRNAs in the biogenesis of trans‐acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21, 798–804. - PubMed
-
- Baeg, K. , Iwakawa, H.O. and Tomari, Y. (2017) The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat. Plants 3, 17036. - PubMed
-
- Béclin, C. , Boutet, S. , Waterhouse, P. and Vaucheret, H. (2002) A branched pathway for transgene‐induced RNA silencing in plants. Curr. Biol. 12, 684–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

