The HCMV Assembly Compartment Is a Dynamic Golgi-Derived MTOC that Controls Nuclear Rotation and Virus Spread
- PMID: 29634939
- PMCID: PMC5896778
- DOI: 10.1016/j.devcel.2018.03.010
The HCMV Assembly Compartment Is a Dynamic Golgi-Derived MTOC that Controls Nuclear Rotation and Virus Spread
Abstract
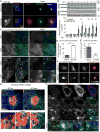
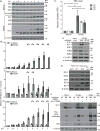
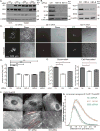
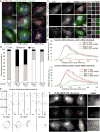
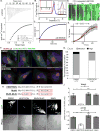
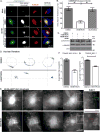
Human cytomegalovirus (HCMV), a leading cause of congenital birth defects, forms an unusual cytoplasmic virion maturation site termed the "assembly compartment" (AC). Here, we show that the AC also acts as a microtubule-organizing center (MTOC) wherein centrosome activity is suppressed and Golgi-based microtubule (MT) nucleation is enhanced. This involved viral manipulation of discrete functions of MT plus-end-binding (EB) proteins. In particular, EB3, but not EB1 or EB2, was recruited to the AC and was required to nucleate MTs that were rapidly acetylated. EB3-regulated acetylated MTs were necessary for nuclear rotation prior to cell migration, maintenance of AC structure, and optimal virus replication. Independently, a myristoylated peptide that blocked EB3-mediated enrichment of MT regulatory proteins at Golgi regions of the AC also suppressed acetylated MT formation, nuclear rotation, and infection. Thus, HCMV offers new insights into the regulation and functions of Golgi-derived MTs and the therapeutic potential of targeting EB3.
Keywords: Golgi; acetylation; cell migration; cytomegalovirus; end-binding protein; microtubule organizing center; nuclear rotation; virus infection.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Y.K. and D.W. filed an invention disclosure on the use of HEBTRON as an antiviral. The authors declare no competing financial interests.
Figures

Comment in
-
HCMV Assembly Is Totally Tubular.Dev Cell. 2018 Apr 9;45(1):1-2. doi: 10.1016/j.devcel.2018.03.014. Dev Cell. 2018. PMID: 29634930
Similar articles
-
Human Cytomegalovirus Exploits TACC3 To Control Microtubule Dynamics and Late Stages of Infection.J Virol. 2021 Aug 25;95(18):e0082121. doi: 10.1128/JVI.00821-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34191581 Free PMC article.
-
Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex.J Virol. 2014 Aug;88(16):9086-99. doi: 10.1128/JVI.01141-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899189 Free PMC article.
-
HCMV Assembly Is Totally Tubular.Dev Cell. 2018 Apr 9;45(1):1-2. doi: 10.1016/j.devcel.2018.03.014. Dev Cell. 2018. PMID: 29634930
-
Microtubule network asymmetry in motile cells: role of Golgi-derived array.Cell Cycle. 2009 Jul 15;8(14):2168-74. doi: 10.4161/cc.8.14.9074. Epub 2009 Jul 20. Cell Cycle. 2009. PMID: 19556895 Free PMC article. Review.
-
Golgi as an MTOC: making microtubules for its own good.Histochem Cell Biol. 2013 Sep;140(3):361-7. doi: 10.1007/s00418-013-1119-4. Epub 2013 Jul 3. Histochem Cell Biol. 2013. PMID: 23821162 Free PMC article. Review.
Cited by
-
RACK1 evolved species-specific multifunctionality in translational control through sequence plasticity within a loop domain.J Cell Sci. 2019 Jun 19;132(12):jcs228908. doi: 10.1242/jcs.228908. J Cell Sci. 2019. PMID: 31118235 Free PMC article.
-
Human Cytomegalovirus Exploits TACC3 To Control Microtubule Dynamics and Late Stages of Infection.J Virol. 2021 Aug 25;95(18):e0082121. doi: 10.1128/JVI.00821-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34191581 Free PMC article.
-
Herpes Simplex Virus Organizes Cytoplasmic Membranes To Form a Viral Assembly Center in Neuronal Cells.J Virol. 2020 Sep 15;94(19):e00900-20. doi: 10.1128/JVI.00900-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32699089 Free PMC article.
-
Localization of the WD repeat-containing protein 5 to the Virion Assembly Compartment Facilitates Human Cytomegalovirus Assembly.J Virol. 2021 Mar 25;95(8):e02101-20. doi: 10.1128/JVI.02101-20. Epub 2021 Jan 27. J Virol. 2021. PMID: 33504601 Free PMC article.
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
References
-
- Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16:711–726. - PubMed
-
- Bazellieres E, Massey-Harroche D, Barthelemy-Requin M, Richard F, Arsanto JP, Le Bivic A. Apico-basal elongation requires a drebrin-E-EB3 complex in columnar human epithelial cells. J Cell Sci. 2012;125:919–931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

