Formation of Chromatin Subcompartments by Phase Separation
- PMID: 29628210
- PMCID: PMC6129460
- DOI: 10.1016/j.bpj.2018.03.011
Formation of Chromatin Subcompartments by Phase Separation
Abstract
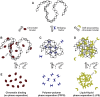
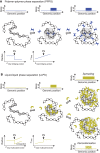
Chromatin is partitioned on multiple length scales into subcompartments that differ from each other with respect to their molecular composition and biological function. It is a key question how these compartments can form even though diffusion constantly mixes the nuclear interior and rapidly balances concentration gradients of soluble nuclear components. Different biophysical concepts are currently used to explain the formation of "chromatin bodies" in a self-organizing manner and without consuming energy. They rationalize how soluble protein factors that are dissolved in the liquid nuclear phase, the nucleoplasm, bind and organize transcriptionally active or silenced chromatin domains. In addition to cooperative binding of proteins to a preformed chromatin structure, two different mechanisms for the formation of phase-separated chromatin subcompartments have been proposed. One is based on bridging proteins that cross-link polymer segments with particular properties. Bridging can induce a collapse of the nucleosome chain and associated factors into an ordered globular phase. The other mechanism is based on multivalent interactions among soluble molecules that bind to chromatin. These interactions can induce liquid-liquid phase separation, which drives the assembly of liquid-like nuclear bodies around the respective binding sites on chromatin. Both phase separation mechanisms can explain that chromatin bodies are dynamic spherical structures, which can coalesce and are in constant and rapid exchange with the surrounding nucleoplasm. However, they make distinct predictions about how the size, density, and stability of chromatin bodies depends on the concentration and interaction behavior of the molecules involved. Here, we compare the different biophysical mechanisms for the assembly of chromatin bodies and discuss experimental strategies to distinguish them from each other. Furthermore, we outline the implications for the establishment and memory of functional chromatin state patterns.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Functional interactions between scaffold proteins, noncoding RNAs, and genome loci induce liquid-liquid phase separation as organizing principle for 3-dimensional nuclear architecture: implications in cancer.FASEB J. 2019 May;33(5):5814-5822. doi: 10.1096/fj.201802715R. Epub 2019 Feb 11. FASEB J. 2019. PMID: 30742773 Review.
-
Biophysical mechanisms of chromatin patterning.Curr Opin Genet Dev. 2020 Apr;61:62-68. doi: 10.1016/j.gde.2020.03.006. Epub 2020 May 11. Curr Opin Genet Dev. 2020. PMID: 32408103 Review.
-
Ephemeral Protein Binding to DNA Shapes Stable Nuclear Bodies and Chromatin Domains.Biophys J. 2017 Mar 28;112(6):1085-1093. doi: 10.1016/j.bpj.2017.01.025. Biophys J. 2017. PMID: 28355537 Free PMC article.
-
Fat nucleosome: Role of lipids on chromatin.Prog Lipid Res. 2018 Apr;70:29-34. doi: 10.1016/j.plipres.2018.04.003. Epub 2018 Apr 17. Prog Lipid Res. 2018. PMID: 29678609 Review.
-
Liquid-Liquid Phase Separation in Chromatin.Cold Spring Harb Perspect Biol. 2022 Feb 1;14(2):a040683. doi: 10.1101/cshperspect.a040683. Cold Spring Harb Perspect Biol. 2022. PMID: 34127447 Free PMC article. Review.
Cited by
-
Super-Resolution Microscopy of Chromatin.Genes (Basel). 2019 Jun 28;10(7):493. doi: 10.3390/genes10070493. Genes (Basel). 2019. PMID: 31261775 Free PMC article. Review.
-
On the relations of phase separation and Hi-C maps to epigenetics.R Soc Open Sci. 2020 Feb 26;7(2):191976. doi: 10.1098/rsos.191976. eCollection 2020 Feb. R Soc Open Sci. 2020. PMID: 32257349 Free PMC article.
-
Biological phase separation: cell biology meets biophysics.Biophys Rev. 2020 Apr;12(2):519-539. doi: 10.1007/s12551-020-00680-x. Epub 2020 Mar 18. Biophys Rev. 2020. PMID: 32189162 Free PMC article. Review.
-
Phasor histone FLIM-FRET microscopy quantifies spatiotemporal rearrangement of chromatin architecture during the DNA damage response.Proc Natl Acad Sci U S A. 2019 Apr 9;116(15):7323-7332. doi: 10.1073/pnas.1814965116. Epub 2019 Mar 27. Proc Natl Acad Sci U S A. 2019. PMID: 30918123 Free PMC article.
-
Integrating transposable elements in the 3D genome.Mob DNA. 2020 Feb 4;11:8. doi: 10.1186/s13100-020-0202-3. eCollection 2020. Mob DNA. 2020. PMID: 32042316 Free PMC article.
References
-
- Rippe K. Dynamic organization of the cell nucleus. Curr. Opin. Genet. Dev. 2007;17:373–380. - PubMed
-
- Cremer T., Cremer M., Cremer C. The 4D nucleome: evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–2943. - PubMed
-
- Boisvert F.M., van Koningsbruggen S., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

