Origin and evolution of the nuclear auxin response system
- PMID: 29580381
- PMCID: PMC5873896
- DOI: 10.7554/eLife.33399
Origin and evolution of the nuclear auxin response system
Abstract
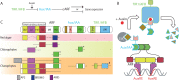
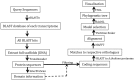
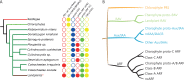
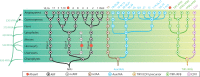
The small signaling molecule auxin controls numerous developmental processes in land plants, acting mostly by regulating gene expression. Auxin response proteins are represented by large families of diverse functions, but neither their origin nor their evolution is understood. Here, we use a deep phylogenomics approach to reconstruct both the origin and the evolutionary trajectory of all nuclear auxin response protein families. We found that, while all subdomains are ancient, a complete auxin response mechanism is limited to land plants. Functional phylogenomics predicts defined steps in the evolution of response system properties, and comparative transcriptomics across six ancient lineages revealed how these innovations shaped a sophisticated response mechanism. Genetic analysis in a basal land plant revealed unexpected contributions of ancient non-canonical proteins in auxin response as well as auxin-unrelated function of core transcription factors. Our study provides a functional evolutionary framework for understanding diverse functions of the auxin signal.
Keywords: A. agrestis; C. richardii; M. polymorpha; P. patens; auxin; charophytes; evolutionary biology; genomics; plant biology.
© 2018, Mutte et al.
Conflict of interest statement
SM, HK, CR, MM, GW, DW No competing interests declared
Figures




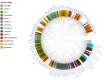

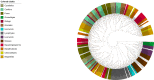
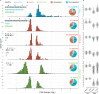
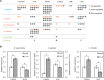


Similar articles
-
Genetic dissection of the auxin response network.Nat Plants. 2020 Sep;6(9):1082-1090. doi: 10.1038/s41477-020-0739-7. Epub 2020 Aug 17. Nat Plants. 2020. PMID: 32807951 Review.
-
Structural Biology of Nuclear Auxin Action.Trends Plant Sci. 2016 Apr;21(4):302-316. doi: 10.1016/j.tplants.2015.10.019. Epub 2015 Dec 1. Trends Plant Sci. 2016. PMID: 26651917 Review.
-
Refining the nuclear auxin response pathway through structural biology.Curr Opin Plant Biol. 2015 Oct;27:22-8. doi: 10.1016/j.pbi.2015.05.007. Epub 2015 Jun 3. Curr Opin Plant Biol. 2015. PMID: 26048079 Free PMC article. Review.
-
Evolution of nuclear auxin signaling: lessons from genetic studies with basal land plants.J Exp Bot. 2018 Jan 4;69(2):291-301. doi: 10.1093/jxb/erx267. J Exp Bot. 2018. PMID: 28992186 Review.
-
Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in Marchantia polymorpha.PLoS Genet. 2015 May 28;11(5):e1005084. doi: 10.1371/journal.pgen.1005084. eCollection 2015 May. PLoS Genet. 2015. PMID: 26020919 Free PMC article.
Cited by
-
A Year at the Forefront of Streptophyte Algal Evolution.Biol Open. 2024 Sep 15;13(9):bio061673. doi: 10.1242/bio.061673. Epub 2024 Sep 19. Biol Open. 2024. PMID: 39297435 Free PMC article. Review.
-
The differential binding and biological efficacy of auxin herbicides.Pest Manag Sci. 2023 Apr;79(4):1305-1315. doi: 10.1002/ps.7294. Epub 2022 Dec 16. Pest Manag Sci. 2023. PMID: 36458868 Free PMC article.
-
Auxin Function in the Brown Alga Dictyota dichotoma.Plant Physiol. 2019 Jan;179(1):280-299. doi: 10.1104/pp.18.01041. Epub 2018 Nov 12. Plant Physiol. 2019. PMID: 30420566 Free PMC article.
-
The birth of a giant: evolutionary insights into the origin of auxin responses in plants.EMBO J. 2023 Mar 15;42(6):e113018. doi: 10.15252/embj.2022113018. Epub 2023 Feb 14. EMBO J. 2023. PMID: 36786017 Free PMC article. Review.
-
An ancestral function of strigolactones as symbiotic rhizosphere signals.Nat Commun. 2022 Jul 8;13(1):3974. doi: 10.1038/s41467-022-31708-3. Nat Commun. 2022. PMID: 35803942 Free PMC article.
References
-
- Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, Morales RL, Berthiaume CT, Parker MS, Djunaedi B, Ingalls AE, Parsek MR, Moran MA, Armbrust EV. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. - DOI - PubMed
-
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, Weijers D, Coll M. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

