Comparative analysis of nanobody sequence and structure data
- PMID: 29569425
- PMCID: PMC6033041
- DOI: 10.1002/prot.25497
Comparative analysis of nanobody sequence and structure data
Abstract
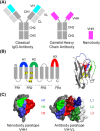
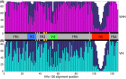
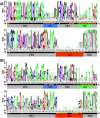
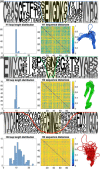
Nanobodies are a class of antigen-binding protein derived from camelids that achieve comparable binding affinities and specificities to classical antibodies, despite comprising only a single 15 kDa variable domain. Their reduced size makes them an exciting target molecule with which we can explore the molecular code that underpins binding specificity-how is such high specificity achieved? Here, we use a novel dataset of 90 nonredundant, protein-binding nanobodies with antigen-bound crystal structures to address this question. To provide a baseline for comparison we construct an analogous set of classical antibodies, allowing us to probe how nanobodies achieve high specificity binding with a dramatically reduced sequence space. Our analysis reveals that nanobodies do not diversify their framework region to compensate for the loss of the VL domain. In addition to the previously reported increase in H3 loop length, we find that nanobodies create diversity by drawing their paratope regions from a significantly larger set of aligned sequence positions, and by exhibiting greater structural variation in their H1 and H2 loops.
Keywords: HcAb; VH; VHH; antibody; camelid; framework; heavy chain antibody; loop; single domain antibody.
© 2018 The Authors Proteins: Structure, Function, and Bioinformatics Published by Wiley Periodicals, Inc.
Figures


Similar articles
-
Analysis of nanobody paratopes reveals greater diversity than classical antibodies.Protein Eng Des Sel. 2018 Jul 1;31(7-8):267-275. doi: 10.1093/protein/gzy017. Protein Eng Des Sel. 2018. PMID: 30053276 Free PMC article.
-
Introduction to heavy chain antibodies and derived Nanobodies.Methods Mol Biol. 2012;911:15-26. doi: 10.1007/978-1-61779-968-6_2. Methods Mol Biol. 2012. PMID: 22886243 Review.
-
NanoBERTa-ASP: predicting nanobody paratope based on a pretrained RoBERTa model.BMC Bioinformatics. 2024 Mar 21;25(1):122. doi: 10.1186/s12859-024-05750-5. BMC Bioinformatics. 2024. PMID: 38515052 Free PMC article.
-
General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold.J Biol Chem. 2009 Jan 30;284(5):3273-3284. doi: 10.1074/jbc.M806889200. Epub 2008 Nov 14. J Biol Chem. 2009. PMID: 19010777
-
Antigen recognition by single-domain antibodies: structural latitudes and constraints.MAbs. 2018 Aug/Sep;10(6):815-826. doi: 10.1080/19420862.2018.1489633. Epub 2018 Aug 15. MAbs. 2018. PMID: 29916758 Free PMC article. Review.
Cited by
-
3D Visualization of Human Blood Vascular Networks Using Single-Domain Antibodies Directed against Endothelial Cell-Selective Adhesion Molecule (ESAM).Int J Mol Sci. 2022 Apr 15;23(8):4369. doi: 10.3390/ijms23084369. Int J Mol Sci. 2022. PMID: 35457187 Free PMC article.
-
Thermodynamic analysis of an entropically driven, high-affinity nanobody-HIV p24 interaction.Biophys J. 2023 Jan 17;122(2):279-289. doi: 10.1016/j.bpj.2022.12.019. Epub 2022 Dec 16. Biophys J. 2023. PMID: 36527237 Free PMC article.
-
Analysis of nanobody paratopes reveals greater diversity than classical antibodies.Protein Eng Des Sel. 2018 Jul 1;31(7-8):267-275. doi: 10.1093/protein/gzy017. Protein Eng Des Sel. 2018. PMID: 30053276 Free PMC article.
-
General Trends of the Camelidae Antibody VHHs Domain Dynamics.Int J Mol Sci. 2023 Feb 24;24(5):4511. doi: 10.3390/ijms24054511. Int J Mol Sci. 2023. PMID: 36901942 Free PMC article.
-
INDI-integrated nanobody database for immunoinformatics.Nucleic Acids Res. 2022 Jan 7;50(D1):D1273-D1281. doi: 10.1093/nar/gkab1021. Nucleic Acids Res. 2022. PMID: 34747487 Free PMC article.
References
-
- Schroeder HW. Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev Comp Immunol. 2006;30(1–2):119–135. - PubMed
-
- Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single‐domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem Sci. 2001;26(4):230–235. - PubMed
-
- Apostoaei AI, Trabalka JR. Review, Synthesis, and Application of Information on the Human Lymphatic System to Radiation Dosimetry for Chronic Lymphocytic Leukemia. SENES Oak Ridge, Oak Ridge, Tennessee, March. [SRDB Ref ID: 119510]; 2012.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

