HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope
- PMID: 29549260
- PMCID: PMC5856820
- DOI: 10.1038/s41467-018-03565-6
HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope
Abstract
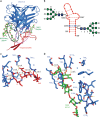
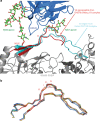
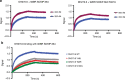
HIV-1 envelope (Env) mimetics are candidate components of prophylactic vaccines and potential therapeutics. Here we use a synthetic V3-glycopeptide ("Man9-V3") for structural studies of an HIV Env third variable loop (V3)-glycan directed, broadly neutralizing antibody (bnAb) lineage ("DH270"), to visualize the epitope on Env and to study how affinity maturation of the lineage proceeded. Unlike many previous V3 mimetics, Man9-V3 encompasses two key features of the V3 region recognized by V3-glycan bnAbs-the conserved GDIR motif and the N332 glycan. In our structure of an antibody fragment of a lineage member, DH270.6, in complex with the V3 glycopeptide, the conformation of the antibody-bound glycopeptide conforms closely to that of the corresponding segment in an intact HIV-1 Env trimer. An additional structure identifies roles for two critical mutations in the development of breadth. The results suggest a strategy for use of a V3 glycopeptide as a vaccine immunogen.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Cooperation between Strain-Specific and Broadly Neutralizing Responses Limited Viral Escape and Prolonged the Exposure of the Broadly Neutralizing Epitope.J Virol. 2017 Aug 24;91(18):e00828-17. doi: 10.1128/JVI.00828-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28679760 Free PMC article.
-
Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide.Sci Transl Med. 2017 Mar 15;9(381):eaai7521. doi: 10.1126/scitranslmed.aai7521. Sci Transl Med. 2017. PMID: 28298421 Free PMC article.
-
A Bispecific Antibody That Simultaneously Recognizes the V2- and V3-Glycan Epitopes of the HIV-1 Envelope Glycoprotein Is Broader and More Potent than Its Parental Antibodies.mBio. 2020 Jan 14;11(1):e03080-19. doi: 10.1128/mBio.03080-19. mBio. 2020. PMID: 31937648 Free PMC article.
-
Envelope proteins of two HIV-1 clades induced different epitope-specific antibody response.Vaccine. 2018 Mar 14;36(12):1627-1636. doi: 10.1016/j.vaccine.2018.01.081. Epub 2018 Feb 9. Vaccine. 2018. PMID: 29429810
-
Vaccine Elicitation of High Mannose-Dependent Neutralizing Antibodies against the V3-Glycan Broadly Neutralizing Epitope in Nonhuman Primates.Cell Rep. 2017 Feb 28;18(9):2175-2188. doi: 10.1016/j.celrep.2017.02.003. Cell Rep. 2017. PMID: 28249163 Free PMC article.
Cited by
-
Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.Cell. 2021 May 27;184(11):2955-2972.e25. doi: 10.1016/j.cell.2021.04.042. Epub 2021 May 20. Cell. 2021. PMID: 34019795 Free PMC article.
-
Structure-Based Stabilization of SOSIP Env Enhances Recombinant Ectodomain Durability and Yield.J Virol. 2023 Jan 31;97(1):e0167322. doi: 10.1128/jvi.01673-22. Epub 2023 Jan 12. J Virol. 2023. PMID: 36633409 Free PMC article.
-
Synthesis and Structure Optimization of Star Copolymers as Tunable Macromolecular Carriers for Minimal Immunogen Vaccine Delivery.Bioconjug Chem. 2024 Aug 21;35(8):1218-1232. doi: 10.1021/acs.bioconjchem.4c00273. Epub 2024 Jul 31. Bioconjug Chem. 2024. PMID: 39081220 Free PMC article.
-
Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.Nat Commun. 2023 May 15;14(1):2782. doi: 10.1038/s41467-023-38108-1. Nat Commun. 2023. PMID: 37188681 Free PMC article.
-
Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers.Retrovirology. 2018 Sep 5;15(1):61. doi: 10.1186/s12977-018-0443-0. Retrovirology. 2018. PMID: 30185183 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

