MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity
- PMID: 29507189
- PMCID: PMC5866576
- DOI: 10.1073/pnas.1718769115
MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity
Abstract
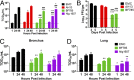
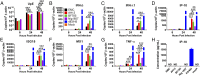
Middle East respiratory syndrome coronavirus (MERS-CoV) causes a zoonotic respiratory disease of global public health concern, and dromedary camels are the only proven source of zoonotic infection. Although MERS-CoV infection is ubiquitous in dromedaries across Africa as well as in the Arabian Peninsula, zoonotic disease appears confined to the Arabian Peninsula. MERS-CoVs from Africa have hitherto been poorly studied. We genetically and phenotypically characterized MERS-CoV from dromedaries sampled in Morocco, Burkina Faso, Nigeria, and Ethiopia. Viruses from Africa (clade C) are phylogenetically distinct from contemporary viruses from the Arabian Peninsula (clades A and B) but remain antigenically similar in microneutralization tests. Viruses from West (Nigeria, Burkina Faso) and North (Morocco) Africa form a subclade, C1, that shares clade-defining genetic signatures including deletions in the accessory gene ORF4b Compared with human and camel MERS-CoV from Saudi Arabia, virus isolates from Burkina Faso (BF785) and Nigeria (Nig1657) had lower virus replication competence in Calu-3 cells and in ex vivo cultures of human bronchus and lung. BF785 replicated to lower titer in lungs of human DPP4-transduced mice. A reverse genetics-derived recombinant MERS-CoV (EMC) lacking ORF4b elicited higher type I and III IFN responses than the isogenic EMC virus in Calu-3 cells. However, ORF4b deletions may not be the major determinant of the reduced replication competence of BF785 and Nig1657. Genetic and phenotypic differences in West African viruses may be relevant to zoonotic potential. There is an urgent need for studies of MERS-CoV at the animal-human interface.
Keywords: Africa; MERS; coronavirus; evolution; zoonosis.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

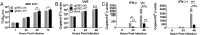
Similar articles
-
Phenotypic and genetic characterization of MERS coronaviruses from Africa to understand their zoonotic potential.Proc Natl Acad Sci U S A. 2021 Jun 22;118(25):e2103984118. doi: 10.1073/pnas.2103984118. Proc Natl Acad Sci U S A. 2021. PMID: 34099577 Free PMC article.
-
Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study.Lancet Respir Med. 2014 Oct;2(10):813-22. doi: 10.1016/S2213-2600(14)70158-4. Epub 2014 Aug 28. Lancet Respir Med. 2014. PMID: 25174549 Free PMC article.
-
Genetic diversity and molecular epidemiology of Middle East Respiratory Syndrome Coronavirus in dromedaries in Ethiopia, 2017-2020.Emerg Microbes Infect. 2023 Dec;12(1):e2164218. doi: 10.1080/22221751.2022.2164218. Emerg Microbes Infect. 2023. PMID: 36620913 Free PMC article.
-
Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction.Pathog Glob Health. 2015;109(8):354-62. doi: 10.1080/20477724.2015.1122852. Pathog Glob Health. 2015. PMID: 26924345 Free PMC article. Review.
-
Dromedary Camels and the Transmission of Middle East Respiratory Syndrome Coronavirus (MERS-CoV).Transbound Emerg Dis. 2017 Apr;64(2):344-353. doi: 10.1111/tbed.12401. Epub 2015 Aug 10. Transbound Emerg Dis. 2017. PMID: 26256102 Free PMC article. Review.
Cited by
-
The impact and future of artificial intelligence in medical genetics and molecular medicine: an ongoing revolution.Funct Integr Genomics. 2024 Aug 16;24(4):138. doi: 10.1007/s10142-024-01417-9. Funct Integr Genomics. 2024. PMID: 39147901 Review.
-
Genomic Diversity and Recombination Analysis of the Spike Protein Gene from Selected Human Coronaviruses.Biology (Basel). 2024 Apr 22;13(4):282. doi: 10.3390/biology13040282. Biology (Basel). 2024. PMID: 38666894 Free PMC article.
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.bioRxiv [Preprint]. 2024 Jun 14:2024.02.27.582110. doi: 10.1101/2024.02.27.582110. bioRxiv. 2024. PMID: 38464327 Free PMC article. Preprint.
-
Prevalence and Molecular Epidemiology of Human Coronaviruses in Africa Prior to the SARS-CoV-2 Outbreak: A Systematic Review.Viruses. 2023 Oct 25;15(11):2146. doi: 10.3390/v15112146. Viruses. 2023. PMID: 38005824 Free PMC article. Review.
-
Contrasting roles of MERS-CoV and SARS-CoV-2 internal proteins in pathogenesis in mice.mBio. 2023 Dec 19;14(6):e0247623. doi: 10.1128/mbio.02476-23. Epub 2023 Oct 26. mBio. 2023. PMID: 37882568 Free PMC article.
References
-
- World Health Organization December 5, 2016 WHO MERS-CoV Global Summary and risk assessment. Available at www.who.int/emergencies/mers-cov/mers-summary-2016.pdf?ua=1. Accessed February 18, 2018.
-
- Al-Tawfiq JA, Perl TM. Middle East respiratory syndrome coronavirus in healthcare settings. Curr Opin Infect Dis. 2015;28:392–396. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

