Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease
- PMID: 29499164
- PMCID: PMC5985341
- DOI: 10.1016/j.ajhg.2018.02.001
Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease
Abstract
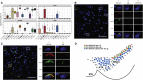
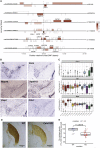
Genetic variation modulating risk of sporadic Parkinson disease (PD) has been primarily explored through genome-wide association studies (GWASs). However, like many other common genetic diseases, the impacted genes remain largely unknown. Here, we used single-cell RNA-seq to characterize dopaminergic (DA) neuron populations in the mouse brain at embryonic and early postnatal time points. These data facilitated unbiased identification of DA neuron subpopulations through their unique transcriptional profiles, including a postnatal neuroblast population and substantia nigra (SN) DA neurons. We use these population-specific data to develop a scoring system to prioritize candidate genes in all 49 GWAS intervals implicated in PD risk, including genes with known PD associations and many with extensive supporting literature. As proof of principle, we confirm that the nigrostriatal pathway is compromised in Cplx1-null mice. Ultimately, this systematic approach establishes biologically pertinent candidates and testable hypotheses for sporadic PD, informing a new era of PD genetic research.
Keywords: Complexin 1; Parkinson disease; dopaminergic neurons; gene regulatory networks; genome-wide association studies; mouse; single-cell RNA sequencing; substantia nigra.
Copyright © 2018 American Society of Human Genetics. All rights reserved.
Figures

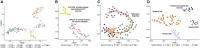

Similar articles
-
The new line of genetically modified mice with constitutive knockout of the gene alpha synuclein to study pathogenetic aspects of differential loss of dopaminergic neurons.Patol Fiziol Eksp Ter. 2016 Jul-Sep;60(3):4-9. Patol Fiziol Eksp Ter. 2016. PMID: 29243900
-
Progressive dopaminergic cell loss with unilateral-to-bilateral progression in a genetic model of Parkinson disease.Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15918-23. doi: 10.1073/pnas.1205102109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23019375 Free PMC article.
-
Repulsive Guidance Molecule a (RGMa) Induces Neuropathological and Behavioral Changes That Closely Resemble Parkinson's Disease.J Neurosci. 2017 Sep 27;37(39):9361-9379. doi: 10.1523/JNEUROSCI.0084-17.2017. Epub 2017 Aug 21. J Neurosci. 2017. PMID: 28842419 Free PMC article.
-
Converging roles of ion channels, calcium, metabolic stress, and activity pattern of Substantia nigra dopaminergic neurons in health and Parkinson's disease.J Neurochem. 2016 Oct;139 Suppl 1(Suppl Suppl 1):156-178. doi: 10.1111/jnc.13572. Epub 2016 Mar 23. J Neurochem. 2016. PMID: 26865375 Free PMC article. Review.
-
Reprint of: revisiting oxidative stress and mitochondrial dysfunction in the pathogenesis of Parkinson disease-resemblance to the effect of amphetamine drugs of abuse.Free Radic Biol Med. 2013 Sep;62:186-201. doi: 10.1016/j.freeradbiomed.2013.05.042. Epub 2013 Jun 3. Free Radic Biol Med. 2013. PMID: 23743292 Review.
Cited by
-
Biological and Medical Importance of Cellular Heterogeneity Deciphered by Single-Cell RNA Sequencing.Cells. 2020 Jul 22;9(8):1751. doi: 10.3390/cells9081751. Cells. 2020. PMID: 32707839 Free PMC article. Review.
-
Gene Families With Stochastic Exclusive Gene Choice Underlie Cell Adhesion in Mammalian Cells.Front Cell Dev Biol. 2021 Apr 29;9:642212. doi: 10.3389/fcell.2021.642212. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33996799 Free PMC article.
-
The molecular landscape of neurological disorders: insights from single-cell RNA sequencing in neurology and neurosurgery.Eur J Med Res. 2023 Nov 16;28(1):529. doi: 10.1186/s40001-023-01504-w. Eur J Med Res. 2023. PMID: 37974227 Free PMC article. Review.
-
Single-cell transcriptional and functional analysis of dopaminergic neurons in organoid-like cultures derived from human fetal midbrain.Development. 2022 Dec 1;149(23):dev200504. doi: 10.1242/dev.200504. Epub 2022 Dec 8. Development. 2022. PMID: 36305490 Free PMC article.
-
Development, wiring and function of dopamine neuron subtypes.Nat Rev Neurosci. 2023 Mar;24(3):134-152. doi: 10.1038/s41583-022-00669-3. Epub 2023 Jan 18. Nat Rev Neurosci. 2023. PMID: 36653531 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

