SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer
- PMID: 29491006
- PMCID: PMC5934778
- DOI: 10.15252/embr.201745124
SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer
Abstract
Peroxisomes account for ~35% of total H2O2 generation in mammalian tissues. Peroxisomal ACOX1 (acyl-CoA oxidase 1) is the first and rate-limiting enzyme in fatty acid β-oxidation and a major producer of H2O2 ACOX1 dysfunction is linked to peroxisomal disorders and hepatocarcinogenesis. Here, we show that the deacetylase sirtuin 5 (SIRT5) is present in peroxisomes and that ACOX1 is a physiological substrate of SIRT5. Mechanistically, SIRT5-mediated desuccinylation inhibits ACOX1 activity by suppressing its active dimer formation in both cultured cells and mouse livers. Deletion of SIRT5 increases H2O2 production and oxidative DNA damage, which can be alleviated by ACOX1 knockdown. We show that SIRT5 downregulation is associated with increased succinylation and activity of ACOX1 and oxidative DNA damage response in hepatocellular carcinoma (HCC). Our study reveals a novel role of SIRT5 in inhibiting peroxisome-induced oxidative stress, in liver protection, and in suppressing HCC development.
Keywords: ACOX1; SIRT5; liver cancer; oxidative stress; succinylation.
© 2018 The Authors.
Figures
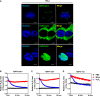
- A
HyPer‐pero, HyPer‐cyto, and HyPer‐nuc were ectopically expressed in HeLa cells, and their subcellular localization was determined by immunofluorescence staining. Representative immunofluorescence images (original magnification, 630×; a single focal plane, scale bar, 5 μm) are shown.
- B–D
HEK293 cells overexpressing the Hyper biosensor were treated with PBS, 500 μM H2O2, or 50 μM menadione for the indicated periods. The H2O2 level in the peroxisome (B), cytosol (C), and nucleus (D) was monitored as described in Materials and Methods.
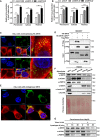
- A, B
Knockdown of SIRT5 stimulates H2O2 production in the peroxisome, cytosol, and nucleus. In Huh7 and HepG2 stable cells with SIRT5 knockdown, endogenous H2O2 production in the indicated cellular compartments was determined by using the Hyper biosensor as described in Materials and Methods. Note: Given that the level of endogenous H2O2 does not change over time (within 30 min, data not shown), we have collected the excitation ratio (490/420 nm) at single time point (at 5 min). Shown are average values with standard deviation (SD) of triplicated experiments. **P < 0.01, and ***P < 0.001 for the indicated comparisons by two‐tailed unpaired Student's t‐test.
- C
Wild‐type SIRT5, but not SIRT5 LQIVdel mutant, co‐localizes with peroxisomal membrane protein PMP70. Immunofluorescence staining was performed in HeLa cells overexpressing HA‐tagged wild‐type or mutant SIRT5 using the indicated antibodies as described in Materials and Methods. Representative immunofluorescence images (original magnification, 630×; a single focal plane, scale bar, 5 μm) are shown.
- D
PEX7 interacts with SIRT5, but not SIRT5 LQIVdel mutant. Flag‐PEX7 was co‐expressed with HA‐tagged wild‐type or mutant SIRT5 in HEK293T cells. Proteins were purified by immunoprecipitation (IP) with Flag beads, followed by Western blot to detect SIRT5 or SIRT5 LQIVdel mutant with an HA antibody.
- E
Endogenous SIRT5 co‐localizes with peroxisomal membrane protein PMP70 in cells. Immunofluorescence staining was performed in HeLa cells using the indicated antibodies as described in Materials and Methods. Representative immunofluorescence images (original magnification, 630×; a single focal plane, scale bar, 5 μm) are shown.
- F
SIRT5 distributes in the peroxisomal fraction. Cellular fractionation was conducted in HepG2 cells to isolate peroxisomes and mitochondria as described in Materials and Methods. The indicated proteins were determined by Western blot analysis. #1 and #2 refer to two repeats from single fractionation.
- G
SIRT5 is a matrix protein rather than a membrane protein in peroxisomes. Isolated peroxisomes from HepG2 cells were subjected to proteinase K (100 μg/ml) digestion with or without 1% Triton X‐100 pre‐treatment as indicated. Harvest the samples at indicated time points to perform Western blot assay. The indicated proteins were determined by Western blot analysis.

The amino acid sequence of SIRT5 protein from the indicated species was imported into “PTSs predictor” of PEROXISOME database (PeroxisomeDB). As shown, SIRT5 contains a predicted PTS2 sequence (marked in red).
SIRT5 interacts with PEX7, but not PEX5. HA‐SIRT5 was co‐expressed with Flag‐PEX5 or Flag‐PEX7 in HEK293T cells. Proteins were purified by immunoprecipitation (IP) with Flag beads, followed by Western blot to detect PEX5 or PEX7 with an HA antibody.
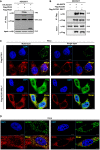
ACOX1 interacts with PEX5. HA‐ACOX1 was co‐overexpressed with Flag‐PEX5 or Flag‐PEX7 in HEK293T cells. PEX proteins were purified by IP with Flag beads, followed by Western blot to detect ACOX1 with an HA antibody.
Both ACOX1 and ACOX1 SKLdel interact with SIRT5. HA‐SIRT5 was co‐overexpressed with Flag‐ACOX1 or Flag‐ACOX1 SKLdel in HEK293T cells, followed by IP with Flag beads and Western blot to detect SIRT5 with an HA antibody.
ACOX1 but not ACOX1 SKLdel mutant is localized in peroxisomes. Flag‐tagged wild‐type or mutant ACOX1 was transiently overexpressed in HeLa cells. Immunofluorescence assay was performed to detect the indicated proteins as described in Materials and Methods. Representative immunofluorescence images (original magnification, 630×; a single focal plane, scale bar, 5 μm) are shown.
ACOX1 co‐localizes with the peroxisomal marker PMP70, but not the mitochondrial marker SDHA. Flag‐ACOX1 was transiently overexpressed in HeLa cells. Immunofluorescence assay was performed to detect the indicated proteins as described in Materials and Methods. Representative immunofluorescence images (original magnification, 630×; a single focal plane, scale bar, 5 μm) are shown.
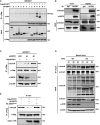
Ectopically expressed ACOX1 interacts with SIRT5. Flag‐ACOX1 was overexpressed in HEK293T cells together with the individual HA‐tagged SIRT proteins as indicated. Proteins were purified by IP with Flag beads, following Western blot to detect SIRT proteins with an HA antibody.
Endogenous ACOX1 interacts with SIRT5. SIRT5 protein in Huh7 and HepG2 liver cells was purified by IP with an anti‐ACOX1 antibody, followed by Western blot to detect SIRT5 with an anti‐SIRT5 antibody.
Knockdown of SIRT5 increases ACOX1 succinylation. In stable HEK293T cells with SIRT5 knockdown, Flag‐ACOX1 was overexpressed. ACOX1 protein was purified by IP with Flag beads and Western blot to detect its succinylation level.
Knockout of Sirt5 increases Acox1 succinylation in mouse livers. Acox1 protein in the liver of Sirt5 KO and wild‐type littermates (n = 3 per group) was purified by IP with an anti‐ACOX1 antibody. ACOX1 succinylation was determined by anti‐succinyl‐lysine antibody.
SIRT5 dessucinylates ACOX1 in vitro. Flag‐ACOX1, HA‐tagged wild‐type SIRT5 and a catalytic inactive mutant SIRT5H158Y were separately overexpressed in HEK293T cells. Proteins were purified by IP with Flag or HA beads, and then eluted with Flag or HA peptide. After incubation with wild‐type or mutant SIRT5 in vitro, the succinylation level of ACOX1 was determined by Western blot analysis.

- A
SIRT5 inactivates ACOX1 in vitro. Flag‐ACOX1, HA‐tagged wild‐type SIRT5, and a catalytic inactive mutant SIRT5H158Y were separately overexpressed in HEK293T cells. Proteins were purified by IP with Flag or HA beads, and then eluted with Flag or HA peptide. After incubation with wild‐type or mutant SIRT5 in vitro, the enzyme activity of ACOX1 was determined as described in Materials and Methods.
- B
ACOX1 activity is increased by lysine succinylation. Purified Flag‐ACOX1 was incubated with or without succinyl‐CoA (0.1 mM) at 37°C for 30 min, followed by measurement of the enzyme activity of ACOX1 as described in Materials and Methods.
- C
Knockdown of SIRT5 increases ACOX1 activity. In stable HEK293T cells with SIRT5 knockdown, Flag‐ACOX1 was ectopically expressed. ACOX1 protein was purified by IP with Flag beads and then eluted with Flag peptide, followed by measurement of its succinylation level and enzyme activity as described in Materials and Methods. The correlation between ACOX1 succinylation level and its enzyme activity is shown.
- D
Knockout of Sirt5 increases Acox1 activity in mouse livers. Acox1 protein in the liver of Sirt5 KO and wild‐type littermates (n = 3 per group) was purified by IP with an anti‐ACOX1 antibody. Acox1 enzyme activity was measured as described in Materials and Methods.
- E, F
SDHA knockdown leads to increased succinylation and activity of ACOX1. In HepG2 cells with stable SDHA knockdown, ACOX1 protein was purified by IP with an anti‐ACOX1 antibody, followed by measurement of its succinylation level (E) and enzyme activity (F) as described in Materials and Methods.
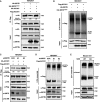
SIRT5 impairs subunit interaction of ACOX1 protein. HA‐tagged wild‐type SIRT5 and a catalytic inactive mutant SIRT5H158Y were each expressed in HEK293T cells co‐expressing HA‐ACOX1 and Flag‐ACOX1. Flag‐ACOX1 was purified by IP with Flag beads, and then, Western blot was performed to detect the interaction with HA‐ACOX1 by an HA antibody.
SIRT5 inhibits the formation of dimeric ACOX1. HA‐tagged wild‐type SIRT5 and a catalytic inactive mutant SIRT5H158Y were each expressed in HEK293T cells co‐expressing Flag‐ACOX1, followed by treatments with or without 2.3% (v/v) glutaraldehyde. The formation of ACOX1 monomer and dimer was determined by Western blotting.
SIRT5 knockdown enhances the interaction of ACOX1 protein subunits. HA‐ACOX1 and Flag‐ACOX1 were transiently co‐expressed in HEK293T cells with stable SIRT5 knockdown. Flag‐ACOX1 was purified by IP with Flag beads, followed by Western blot to detect its interaction with HA‐ACOX1 with an HA antibody.
SIRT5 knockdown promotes the formation of dimeric ACOX1. Flag‐ACOX1 was expressed in HEK293T cells with stable SIRT5 knockdown. The transfected cell lysates were treated with 2.3% (v/v) glutaraldehyde. The formation of ACOX1 monomer and dimer was determined by Western blotting.
Knockout of Sirt5 promotes the formation of dimeric Acox1 in mouse livers. Acox1 protein in the liver of Sirt5 KO and wild‐type littermate was purified by IP with an anti‐ACOX1 antibody, followed by treatment with 2.3% (v/v) glutaraldehyde. The formation of Acox1 monomer and dimer was determined by Western blotting.
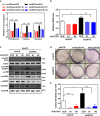
- A
ACOX1 knockdown diminishes the effect of SIRT5 knockdown on increasing H2O2 production. In HepG2 stable cells with single or double knockdown of SIRT5/ACOX1, H2O2 production in the indicated cellular compartments was determined by using the Hyper biosensor as described in Materials and Methods.
- B
ACOX1 knockdown impairs the effect of SIRT5 knockdown on increasing ROS. In HepG2 stable cells with single or double knockdown of SIRT5/ACOX1, ROS level was determined in cell extracts as described in Materials and Methods.
- C
ACOX1 knockdown impairs the effect of SIRT5 knockdown on inducing oxidative DNA damage response. In HepG2 stable cells with single or double knockdown of SIRT5/ACOX1, the indicated classical oxidative DNA damage response markers were determined by Western blot analysis, completed in biological triplicate.
- D, E
ACOX1 knockdown impairs the effect of SIRT5 knockdown on promoting anchorage‐independent growth in HepG2 liver cells. The capability of HepG2 cells with single or double knockdown of SIRT5/ACOX1 to exhibit anchorage‐independent growth was determined by performing soft‐agar colony formation assay as described in Materials and Methods. Representative images for soft‐agar colony formation (D) and the related quantified result (E) are shown. The other images are presented in Appendix Fig S11.
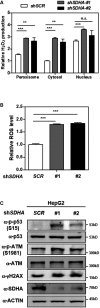
Knockdown of SDHA stimulates H2O2 production in the peroxisome, cytosol, and nucleus. In HepG2 cells with stable SDHA knockdown, endogenous H2O2 production in the indicated cellular compartments was determined by using the Hyper biosensor as described in Materials and Methods. Note: Given that the level of endogenous H2O2 does not change over time, we have collected the excitation ratio (490/420 nm) at single time point (at 5 min).
Knockdown of SDHA increases cellular ROS. In HepG2 cells with stable SDHA knockdown, ROS level was determined in cell extracts as described in Materials and Methods.
Knockdown of SDHA induces oxidative DNA damage response. In HepG2 cells with stable SDHA knockdown, the indicated classical oxidative DNA damage response markers were determined by Western blot analysis.
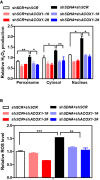
ACOX1 knockdown partially diminishes the effect of SDHA knockdown on increasing H2O2. In HepG2 stable cells with single or double knockdown of SDHA/ACOX1, endogenous H2O2 level in the indicated cellular compartments was determined by using the Hyper biosensor as described in Materials and Methods. Note: given that the level of endogenous H2O2 does not change over time, we have collected the excitation ratio (490/420 nm) at single time point (at 5 min).
ACOX1 knockdown partially impairs the effect of SDHA knockdown on increasing ROS. In HepG2 stable cells with single or double knockdown of SDHA/ACOX1, ROS level was determined in cell extracts as described in Materials and Methods.
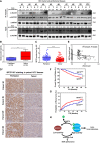
- A
HCC tumors show reduced SIRT5 expression and increased ACOX1 succinylation. In total, 10 pairs of HCC tumor tissues (T) and peritumoral tissues (P) paired samples, ACOX1 protein was immunopurified with an anti‐ACOX1 antibody, followed by Western blot to detect its succinylation. Protein levels of ACOX1 and SIRT5 were determined by direct Western blot. Relative ACOX1 and SIRT5 protein levels were normalized against β‐actin protein.
- B
ACOX1 activity is increased in HCC tumors. Purified ACOX1 protein in 10 pairs of HCC tumors and peritumoral tissues in (A) was subjected to its enzyme activity assay as described in Materials and Methods. The graph boxes delimit the first and third quartiles. The horizontal lines represent the data medians. Whiskers delimit the lowest and the highest value within 1.5 of the interquartile range (IQR) of the lower and the upper quartiles, respectively.
- C, D
SIRT5 protein expression is downregulated in HCC tumors. In a study cohort consisting of 78 HCC patients, the SIRT5 protein was detected by IHC staining as described in Materials and Methods (D). Representative IHC images (original magnification, 200×; a single focal plane, scale bar, 50 μm) are shown.
- E
γH2AX levels show a negative correlation with SIRT5 protein levels in HCC tumors. In a cohort consisting of 118 HCC patients (each has two repeats), the γH2AX and SIRT5 protein levels in the same samples were detected by IHC staining as described in Materials and Methods, followed by quantitative assessment.
- F, G
High SIRT5 expression is associated with favorable prognosis in HCC patients. In a tissue microarray composed of primary tumors from 316 consecutive curative HCC patients, the SIRT5 protein was detected by IHC staining as described in Materials and Methods. The correlation between SIRT5 staining intensity and OS (overall survival) or TTR (time to recurrence) was assessed by Kaplan–Meier method and log‐rank test, respectively.
- H
A proposed model illustrating SIRT5‐dependent lysine succinylation in regulating ACOX1 activity. As shown, lysine succinylation is increased by either SIRT5 loss or SDH dysfunction. Lysine succinylation can activate ACOX1 by promoting the formation of dimeric ACOX1, thereby stimulating H2O2 production in peroxisomes.
Similar articles
-
Evidence of oxidative stress in very long chain fatty acid--treated oligodendrocytes and potentialization of ROS production using RNA interference-directed knockdown of ABCD1 and ACOX1 peroxisomal proteins.Neuroscience. 2012 Jun 28;213:1-18. doi: 10.1016/j.neuroscience.2012.03.058. Epub 2012 Apr 19. Neuroscience. 2012. PMID: 22521832
-
Peroxisomal β-oxidation regulates whole body metabolism, inflammatory vigor, and pathogenesis of nonalcoholic fatty liver disease.JCI Insight. 2018 Mar 22;3(6):e93626. doi: 10.1172/jci.insight.93626. JCI Insight. 2018. PMID: 29563328 Free PMC article.
-
Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function.Proc Natl Acad Sci U S A. 2016 Apr 19;113(16):4320-5. doi: 10.1073/pnas.1519858113. Epub 2016 Apr 5. Proc Natl Acad Sci U S A. 2016. PMID: 27051063 Free PMC article.
-
Hydrogen peroxide generation in peroxisome proliferator-induced oncogenesis.Mutat Res. 2000 Mar 17;448(2):159-77. doi: 10.1016/s0027-5107(99)00234-1. Mutat Res. 2000. PMID: 10725470 Review.
-
Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology.Crit Rev Biochem Mol Biol. 2018 Jun;53(3):311-334. doi: 10.1080/10409238.2018.1458071. Epub 2018 Apr 11. Crit Rev Biochem Mol Biol. 2018. PMID: 29637793 Free PMC article. Review.
Cited by
-
Succinylation Regulators Promote Clear Cell Renal Cell Carcinoma by Immune Regulation and RNA N6-Methyladenosine Methylation.Front Cell Dev Biol. 2021 Feb 18;9:622198. doi: 10.3389/fcell.2021.622198. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33681201 Free PMC article.
-
Metabolite sensing and signaling in cell metabolism.Signal Transduct Target Ther. 2018 Nov 9;3:30. doi: 10.1038/s41392-018-0024-7. eCollection 2018. Signal Transduct Target Ther. 2018. PMID: 30416760 Free PMC article. Review.
-
Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials.Signal Transduct Target Ther. 2019 Dec 17;4:62. doi: 10.1038/s41392-019-0095-0. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 31871779 Free PMC article. Review.
-
Generation of multicellular tumor spheroids with micro-well array for anticancer drug combination screening based on a valuable biomarker of hepatocellular carcinoma.Front Bioeng Biotechnol. 2022 Dec 1;10:1087656. doi: 10.3389/fbioe.2022.1087656. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36532586 Free PMC article.
-
Sirtuins and Insulin Resistance.Front Endocrinol (Lausanne). 2018 Dec 6;9:748. doi: 10.3389/fendo.2018.00748. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 30574122 Free PMC article. Review.
References
-
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK (2006) Peroxisomal beta‐oxidation–a metabolic pathway with multiple functions. Biochem Biophys Acta 1763: 1413–1426 - PubMed
-
- Wanders RJ, Ferdinandusse S, Brites P, Kemp S (2010) Peroxisomes, lipid metabolism and lipotoxicity. Biochem Biophys Acta 1801: 272–280 - PubMed
-
- Shimozawa N (2011) Molecular and clinical findings and diagnostic flowchart of peroxisomal diseases. Brain Develop 33: 770–776 - PubMed
-
- Lazarow PB (2003) Peroxisome biogenesis: advances and conundrums. Curr Opin Cell Biol 15: 489–497 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

