Revealing nascent proteomics in signaling pathways and cell differentiation
- PMID: 29467287
- PMCID: PMC5877968
- DOI: 10.1073/pnas.1707514115
Revealing nascent proteomics in signaling pathways and cell differentiation
Abstract
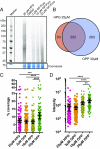
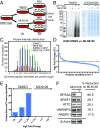
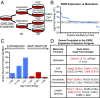
Regulation of gene expression at the level of protein synthesis is a crucial element in driving how the genetic landscape is expressed. However, we are still limited in technologies that can quantitatively capture the immediate proteomic changes that allow cells to respond to specific stimuli. Here, we present a method to capture and identify nascent proteomes in situ across different cell types without disturbing normal growth conditions, using O-propargyl-puromycin (OPP). Cell-permeable OPP rapidly labels nascent elongating polypeptides, which are subsequently conjugated to biotin-azide, using click chemistry, and captured with streptavidin beads, followed by digestion and analysis, using liquid chromatography-tandem mass spectrometry. Our technique of OPP-mediated identification (OPP-ID) allows detection of widespread proteomic changes within a short 2-hour pulse of OPP. We illustrate our technique by recapitulating alterations of proteomic networks induced by a potent mammalian target of rapamycin inhibitor, MLN128. In addition, by employing OPP-ID, we identify more than 2,100 proteins and uncover distinct protein networks underlying early erythroid progenitor and differentiation states not amenable to alternative approaches such as amino acid analog labeling. We present OPP-ID as a method to quantitatively identify nascent proteomes across an array of biological contexts while preserving the subtleties directing signaling in the native cellular environment.
Keywords: erythropoiesis; mTOR; proteomics; puromycin; translation.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
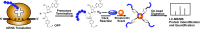
Similar articles
-
Rapid cell type-specific nascent proteome labeling in Drosophila.Elife. 2023 Apr 24;12:e83545. doi: 10.7554/eLife.83545. Elife. 2023. PMID: 37092974 Free PMC article.
-
Quantitative nascent proteome profiling by dual-pulse labelling with O-propargyl-puromycin and stable isotope-labelled amino acids.J Biochem. 2021 Mar 5;169(2):227-236. doi: 10.1093/jb/mvaa104. J Biochem. 2021. PMID: 32926143
-
Capture, Release, and Identification of Newly Synthesized Proteins for Improved Profiling of Functional Translatomes.Mol Cell Proteomics. 2023 Mar;22(3):100497. doi: 10.1016/j.mcpro.2023.100497. Epub 2023 Jan 13. Mol Cell Proteomics. 2023. PMID: 36642223 Free PMC article.
-
Novel approaches to identify protein adducts produced by lipid peroxidation.Free Radic Res. 2015;49(7):881-7. doi: 10.3109/10715762.2015.1019348. Epub 2015 Mar 30. Free Radic Res. 2015. PMID: 25819163 Free PMC article. Review.
-
Quantitative proteome mapping of nitrotyrosines.Methods Enzymol. 2008;440:191-205. doi: 10.1016/S0076-6879(07)00811-7. Methods Enzymol. 2008. PMID: 18423218 Review.
Cited by
-
Focal adhesion-derived liquid-liquid phase separations regulate mRNA translation.bioRxiv [Preprint]. 2024 Nov 14:2023.11.22.568289. doi: 10.1101/2023.11.22.568289. bioRxiv. 2024. PMID: 38045367 Free PMC article. Preprint.
-
Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease.Nat Neurosci. 2019 Mar;22(3):401-412. doi: 10.1038/s41593-018-0332-9. Epub 2019 Feb 11. Nat Neurosci. 2019. PMID: 30742114 Free PMC article.
-
Translatome Regulation in Neuronal Injury and Axon Regrowth.eNeuro. 2018 May 10;5(2):ENEURO.0276-17.2018. doi: 10.1523/ENEURO.0276-17.2018. eCollection 2018 Mar-Apr. eNeuro. 2018. PMID: 29756027 Free PMC article.
-
Ribosomal protein S11 influences glioma response to TOP2 poisons.Oncogene. 2020 Jul;39(27):5068-5081. doi: 10.1038/s41388-020-1342-0. Epub 2020 Jun 11. Oncogene. 2020. PMID: 32528131 Free PMC article.
-
Rapid cell type-specific nascent proteome labeling in Drosophila.Elife. 2023 Apr 24;12:e83545. doi: 10.7554/eLife.83545. Elife. 2023. PMID: 37092974 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

