GRL-079, a Novel HIV-1 Protease Inhibitor, Is Extremely Potent against Multidrug-Resistant HIV-1 Variants and Has a High Genetic Barrier against the Emergence of Resistant Variants
- PMID: 29463535
- PMCID: PMC5923169
- DOI: 10.1128/AAC.02060-17
GRL-079, a Novel HIV-1 Protease Inhibitor, Is Extremely Potent against Multidrug-Resistant HIV-1 Variants and Has a High Genetic Barrier against the Emergence of Resistant Variants
Abstract
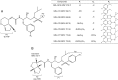
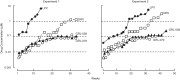
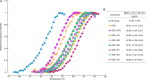
We identified four novel nonpeptidic human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PIs), GRL-078, -079, -077, and -058, containing an alkylamine at the C-5 position of P2 tetrahydropyrano-tetrahydrofuran (Tp-THF) and a P2' cyclopropyl (Cp) (or isopropyl)-aminobenzothiazole (Abt) moiety. Their 50% effective concentrations (EC50s) were 2.5 to 30 nM against wild-type HIV-1NL4-3, 0.3 to 6.7 nM against HIV-2EHO, and 0.9 to 90 nM against laboratory-selected PI-resistant HIV-1 and clinical HIV-1 variants resistant to multiple FDA-approved PIs (HIVMDR). GRL-078, -079, -077, and -058 also effectively blocked the replication of HIV-1 variants highly resistant to darunavir (DRV) (HIVDRVrp51), with EC50s of 38, 62, 61, and 90 nM, respectively, while four FDA-approved PIs examined (amprenavir, atazanavir, lopinavir [LPV], and DRV) had virtually no activity (EC50s of >1,000 nM) against HIVDRVrp51 Structurally, GRL-078, -079, and -058 form strong hydrogen bond interactions between Tp-THF modified at C-5 and Asp29/Asp30/Gly48 of wild-type protease, while the P2' Cp-Abt group forms strong hydrogen bonds with Asp30'. The Tp-THF and Cp-Abt moieties also have good nonpolar interactions with protease residues located in the flap region. For selection with LPV and DRV by use of a mixture of 11 HIVMDR strains (HIV11MIX), HIV11MIX became highly resistant to LPV and DRV over 13 to 32 and 32 to 41 weeks, respectively. However, for selection with GRL-079 and GRL-058, HIV11MIX failed to replicate at >0.08 μM and >0.2 μM, respectively. Thermal stability results supported the highly favorable anti-HIV-1 potency of GRL-079 as well as other PIs. The present data strongly suggest that the P2 Tp-THF group modified at C-5 and the P2' Abt group contribute to the potent anti-HIV-1 profiles of the four PIs against HIV-1NL4-3 and a wide spectrum of HIVMDR strains.
Keywords: AIDS; HIV-1; protease; protease inhibitors.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.J Virol. 2015 Nov 18;90(5):2180-94. doi: 10.1128/JVI.01829-15. J Virol. 2015. PMID: 26581995 Free PMC article.
-
Novel Protease Inhibitors Containing C-5-Modified bis-Tetrahydrofuranylurethane and Aminobenzothiazole as P2 and P2' Ligands That Exert Potent Antiviral Activity against Highly Multidrug-Resistant HIV-1 with a High Genetic Barrier against the Emergence of Drug Resistance.Antimicrob Agents Chemother. 2019 Jul 25;63(8):e00372-19. doi: 10.1128/AAC.00372-19. Print 2019 Aug. Antimicrob Agents Chemother. 2019. PMID: 31085520 Free PMC article.
-
GRL-04810 and GRL-05010, difluoride-containing nonpeptidic HIV-1 protease inhibitors (PIs) that inhibit the replication of multi-PI-resistant HIV-1 in vitro and possess favorable lipophilicity that may allow blood-brain barrier penetration.Antimicrob Agents Chemother. 2013 Dec;57(12):6110-21. doi: 10.1128/AAC.01420-13. Epub 2013 Sep 30. Antimicrob Agents Chemother. 2013. PMID: 24080647 Free PMC article.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
Cited by
-
A novel HIV-1 protease inhibitor, GRL-044, has potent activity against various HIV-1s with an extremely high genetic barrier to the emergence of HIV-1 drug resistance.Glob Health Med. 2019 Oct 31;1(1):36-48. doi: 10.35772/ghm.2019.01003. Glob Health Med. 2019. PMID: 33330753 Free PMC article.
-
Piperidine scaffold as the novel P2-ligands in cyclopropyl-containing HIV-1 protease inhibitors: Structure-based design, synthesis, biological evaluation and docking study.PLoS One. 2020 Jul 22;15(7):e0235483. doi: 10.1371/journal.pone.0235483. eCollection 2020. PLoS One. 2020. PMID: 32697773 Free PMC article.
-
Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance.Chem Commun (Camb). 2022 Oct 20;58(84):11762-11782. doi: 10.1039/d2cc04541a. Chem Commun (Camb). 2022. PMID: 36200462 Free PMC article. Review.
-
Selection of HIV-1 for resistance to fifth-generation protease inhibitors reveals two independent pathways to high-level resistance.Elife. 2023 Mar 15;12:e80328. doi: 10.7554/eLife.80328. Elife. 2023. PMID: 36920025 Free PMC article.
-
Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.Antimicrob Agents Chemother. 2019 May 24;63(6):e02635-18. doi: 10.1128/AAC.02635-18. Print 2019 Jun. Antimicrob Agents Chemother. 2019. PMID: 30962341 Free PMC article.
References
-
- Edmonds A, Yotebieng M, Lusiama J, Matumona Y, Kitetele F, Napravnik S, Cole SR, Van Rie A, Behets F. 2011. The effect of highly active antiretroviral therapy on the survival of HIV-infected children in a resource-deprived setting: a cohort study. PLoS Med 8:e1001044. doi:10.1371/journal.pmed.1001044. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

