Interaction of Human Cytomegalovirus Tegument Proteins ppUL35 and ppUL35A with Sorting Nexin 5 Regulates Glycoprotein B (gpUL55) Localization
- PMID: 29444945
- PMCID: PMC5899209
- DOI: 10.1128/JVI.00013-18
Interaction of Human Cytomegalovirus Tegument Proteins ppUL35 and ppUL35A with Sorting Nexin 5 Regulates Glycoprotein B (gpUL55) Localization
Abstract
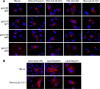
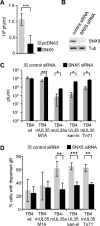
Human cytomegalovirus (HCMV) is a widespread human pathogen that causes asymptomatic infection in healthy individuals but poses a serious threat to immunocompromised patients. During the late phase of HCMV infection, the viral capsid is transported to the cytoplasmic viral assembly center (cVAC), where it is enclosed by the tegument protein layer and the viral envelope. The cVAC consists of circularly arranged vesicles from the trans-Golgi and endosomal networks. The HCMV gene UL35 encodes ppUL35 and its shorter form, ppUL35A. We have previously shown that the UL35 gene is involved in HCMV assembly, but it is unknown how UL35 proteins regulate viral assembly. Here we show that sorting nexin 5 (SNX5), a component of the retromer and part of the retrograde transport pathway, interacts with UL35 proteins. Expression of wild-type proteins but not mutants defective in SNX5 binding resulted in the cellular redistribution of the cation-independent mannose-6-phosphate receptor (CI-M6PR), indicating that UL35 proteins bind and negatively regulate SNX5 to modulate cellular transport pathways. Furthermore, binding of UL35 proteins to SNX5 was required for efficient viral replication and for transport of the most abundant HCMV glycoprotein B (gB; gpUL55) to the cVAC. These results indicate that ppUL35 and ppUL35A control the localization of the essential gB through the regulation of a retrograde transport pathway. Thus, this work is the first to define a molecular interaction between a tegument protein and a vesicular transport factor to regulate glycoprotein localization.IMPORTANCE Human cytomegalovirus is ubiquitously present in the healthy population, but reactivation or reinfection can cause serious, life-threatening infections in immunocompromised patients. For completion of its lytic cycle, human cytomegalovirus induces formation of an assembly center where mature virus particles are formed from multiple viral proteins. Viral glycoproteins use separate vesicular pathways for transport to the assembly center, which are incompletely understood. Our research identified a viral structural protein which affects the localization of one of the major glycoproteins. We could link this change in glycoprotein localization to an interaction of the structural protein with a cellular protein involved in regulation of vesicle transport. This increases our understanding of how the virus intersects into cellular regulatory pathways to enhance its own replication.
Keywords: UL35; cytomegalovirus; glycoprotein B; sorting nexin 5.
Copyright © 2018 American Society for Microbiology.
Figures
Similar articles
-
Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation.J Virol. 2005 Mar;79(5):3084-96. doi: 10.1128/JVI.79.5.3084-3096.2005. J Virol. 2005. PMID: 15709028 Free PMC article.
-
Potent Inhibition of Human Cytomegalovirus by Modulation of Cellular SNARE Syntaxin 5.J Virol. 2016 Dec 16;91(1):e01637-16. doi: 10.1128/JVI.01637-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795424 Free PMC article.
-
Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer.J Virol. 2017 May 12;91(11):e00205-17. doi: 10.1128/JVI.00205-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331097 Free PMC article.
-
Human cytomegalovirus glycoprotein-receptor interactions.Transplant Proc. 1991 Jun;23(3 Suppl 3):60-3. Transplant Proc. 1991. PMID: 1648838 Review.
-
Human cytomegalovirus tegument proteins (pp65, pp71, pp150, pp28).Virol J. 2012 Jan 17;9:22. doi: 10.1186/1743-422X-9-22. Virol J. 2012. PMID: 22251420 Free PMC article. Review.
Cited by
-
The Cytomegalovirus Tegument Protein UL35 Antagonizes Pattern Recognition Receptor-Mediated Type I IFN Transcription.Microorganisms. 2020 May 26;8(6):790. doi: 10.3390/microorganisms8060790. Microorganisms. 2020. PMID: 32466380 Free PMC article.
-
Sorting nexin 5 mediates virus-induced autophagy and immunity.Nature. 2021 Jan;589(7842):456-461. doi: 10.1038/s41586-020-03056-z. Epub 2020 Dec 16. Nature. 2021. PMID: 33328639 Free PMC article.
-
Human Cytomegalovirus Primary Infection and Reactivation: Insights From Virion-Carried Molecules.Front Microbiol. 2020 Jul 14;11:1511. doi: 10.3389/fmicb.2020.01511. eCollection 2020. Front Microbiol. 2020. PMID: 32765441 Free PMC article. Review.
-
Targeting Conserved Sequences Circumvents the Evolution of Resistance in a Viral Gene Drive against Human Cytomegalovirus.J Virol. 2021 Jul 12;95(15):e0080221. doi: 10.1128/JVI.00802-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011551 Free PMC article.
-
Virus-host protein interactions as footprints of human cytomegalovirus replication.Curr Opin Virol. 2022 Feb;52:135-147. doi: 10.1016/j.coviro.2021.11.016. Epub 2021 Dec 16. Curr Opin Virol. 2022. PMID: 34923282 Free PMC article. Review.
References
-
- Winkler M. 2003. Interactions and functions of human cytomegalovirus tegument proteins, p 113–121. In Pr̈osch S, Cinatl J, Scholz M (ed), New aspects of CMV-related immunopathology. Karger, Basel, Switzerland.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

