TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells
- PMID: 29443960
- PMCID: PMC6028240
- DOI: 10.1038/nature25501
TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells
Abstract
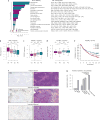
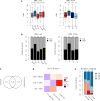
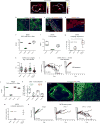
Therapeutic antibodies that block the programmed death-1 (PD-1)-programmed death-ligand 1 (PD-L1) pathway can induce robust and durable responses in patients with various cancers, including metastatic urothelial cancer. However, these responses only occur in a subset of patients. Elucidating the determinants of response and resistance is key to improving outcomes and developing new treatment strategies. Here we examined tumours from a large cohort of patients with metastatic urothelial cancer who were treated with an anti-PD-L1 agent (atezolizumab) and identified major determinants of clinical outcome. Response to treatment was associated with CD8+ T-effector cell phenotype and, to an even greater extent, high neoantigen or tumour mutation burden. Lack of response was associated with a signature of transforming growth factor β (TGFβ) signalling in fibroblasts. This occurred particularly in patients with tumours, which showed exclusion of CD8+ T cells from the tumour parenchyma that were instead found in the fibroblast- and collagen-rich peritumoural stroma; a common phenotype among patients with metastatic urothelial cancer. Using a mouse model that recapitulates this immune-excluded phenotype, we found that therapeutic co-administration of TGFβ-blocking and anti-PD-L1 antibodies reduced TGFβ signalling in stromal cells, facilitated T-cell penetration into the centre of tumours, and provoked vigorous anti-tumour immunity and tumour regression. Integration of these three independent biological features provides the best basis for understanding patient outcome in this setting and suggests that TGFβ shapes the tumour microenvironment to restrain anti-tumour immunity by restricting T-cell infiltration.
Conflict of interest statement
Pontus Eriksson, Mattias Hoglund, Lawrence Fong, Stephen Santoro have no competing interests.
Figures



Comment in
-
TGFβ Promotes Immune Evasion to Limit the Efficacy of Anti-PD-1/PD-L1.Cancer Discov. 2018 Apr;8(4):OF10. doi: 10.1158/2159-8290.CD-RW2018-034. Epub 2018 Feb 23. Cancer Discov. 2018. PMID: 29475883
-
Bladder cancer: Mechanisms of anti-PDL1 resistance.Nat Rev Urol. 2018 Apr;15(4):201. doi: 10.1038/nrurol.2018.28. Epub 2018 Mar 6. Nat Rev Urol. 2018. PMID: 29508844 No abstract available.
-
Immunotherapy: Tear down this wall.Nat Rev Cancer. 2018 Apr;18(4):205. doi: 10.1038/nrc.2018.22. Epub 2018 Mar 9. Nat Rev Cancer. 2018. PMID: 29520089 No abstract available.
-
Immunotherapy: Tear down this wall.Nat Rev Immunol. 2018 Apr;18(4):221. doi: 10.1038/nri.2018.20. Epub 2018 Mar 16. Nat Rev Immunol. 2018. PMID: 29545641 No abstract available.
-
Dual Transforming Growth Factor-β and Programmed Death-1 Blockade: A Strategy for Immune-Excluded Tumors?Trends Immunol. 2018 Jun;39(6):435-437. doi: 10.1016/j.it.2018.03.002. Epub 2018 Mar 26. Trends Immunol. 2018. PMID: 29598848 Free PMC article.
-
TGF-β Inhibition and Immunotherapy: Checkmate.Immunity. 2018 Apr 17;48(4):626-628. doi: 10.1016/j.immuni.2018.03.037. Immunity. 2018. PMID: 29669246 Free PMC article.
Similar articles
-
Anti-PD-L1/TGFβR2 (M7824) fusion protein induces immunogenic modulation of human urothelial carcinoma cell lines, rendering them more susceptible to immune-mediated recognition and lysis.Urol Oncol. 2018 Mar;36(3):93.e1-93.e11. doi: 10.1016/j.urolonc.2017.09.027. Epub 2017 Nov 2. Urol Oncol. 2018. PMID: 29103968 Free PMC article.
-
Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis.PLoS Med. 2017 May 26;14(5):e1002309. doi: 10.1371/journal.pmed.1002309. eCollection 2017 May. PLoS Med. 2017. PMID: 28552987 Free PMC article. Clinical Trial.
-
A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study.Lancet Oncol. 2018 Sep;19(9):1180-1191. doi: 10.1016/S1470-2045(18)30413-3. Epub 2018 Aug 14. Lancet Oncol. 2018. PMID: 30120041
-
A Systematic Review of Immunotherapy in Urologic Cancer: Evolving Roles for Targeting of CTLA-4, PD-1/PD-L1, and HLA-G.Eur Urol. 2015 Aug;68(2):267-79. doi: 10.1016/j.eururo.2015.02.032. Epub 2015 Mar 29. Eur Urol. 2015. PMID: 25824720 Review.
-
Avelumab: A Novel Anti-PD-L1 Agent in the Treatment of Merkel Cell Carcinoma and Urothelial Cell Carcinoma.Crit Rev Immunol. 2018;38(3):159-206. doi: 10.1615/CritRevImmunol.2018025204. Crit Rev Immunol. 2018. PMID: 30004857 Review.
Cited by
-
Study on correlation between CXCL13 and prognosis and immune characteristics of ovarian cancer.Medicine (Baltimore). 2024 Oct 25;103(43):e40272. doi: 10.1097/MD.0000000000040272. Medicine (Baltimore). 2024. PMID: 39470479 Free PMC article.
-
M2 Macrophage Co-Expression Factors Correlate With Immune Phenotype and Predict Prognosis of Bladder Cancer.Front Oncol. 2021 Mar 22;11:609334. doi: 10.3389/fonc.2021.609334. eCollection 2021. Front Oncol. 2021. PMID: 33828973 Free PMC article.
-
Coming of Age of Immunotherapy of Urothelial Cancer.Target Oncol. 2021 May;16(3):283-294. doi: 10.1007/s11523-021-00804-7. Epub 2021 Mar 12. Target Oncol. 2021. PMID: 33710533 Review.
-
Immunosuppressive Microenvironment Revealed by Immune Cell Landscape in Pre-metastatic Liver of Colorectal Cancer.Front Oncol. 2021 Mar 23;11:620688. doi: 10.3389/fonc.2021.620688. eCollection 2021. Front Oncol. 2021. PMID: 33833986 Free PMC article.
-
Secreted gelsolin inhibits DNGR-1-dependent cross-presentation and cancer immunity.Cell. 2021 Jul 22;184(15):4016-4031.e22. doi: 10.1016/j.cell.2021.05.021. Epub 2021 Jun 2. Cell. 2021. PMID: 34081922 Free PMC article.
References
-
- Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HEK, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. - PMC - PubMed
-
- Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng S-L, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. - PubMed
-
- Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee J-L, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, Necchi A, Gerritsen W, Gurney H, Quinn DI, Culine S, Sternberg CN, Mai Y, Poehlein CH, Perini RF, Bajorin DF KEYNOTE-045 Investigators. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med. 2017;376:1015–1026. - PMC - PubMed
-
- Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O'Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. - PMC - PubMed
-
- Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, Dawson NA, van der Heijden MS, Dreicer R, Srinivas S, Retz MM, Joseph RW, Drakaki A, Vaishampayan UN, Sridhar SS, Quinn DI, Durán I, Shaffer DR, Eigl BJ, Grivas PD, Yu EY, Li S, Kadel EE, Boyd Z, Bourgon R, Hegde PS, Mariathasan S, Thåström A, Abidoye OO, Fine GD, Bajorin DF IMvigor210 Study Group. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

