The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation
- PMID: 29415178
- PMCID: PMC5909452
- DOI: 10.1093/nar/gky070
The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation
Abstract
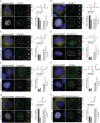
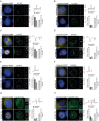
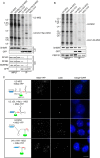
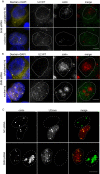
Cajal bodies (CBs) are nuclear non-membrane bound organelles where small nuclear ribonucleoprotein particles (snRNPs) undergo their final maturation and quality control before they are released to the nucleoplasm. However, the molecular mechanism how immature snRNPs are targeted and retained in CBs has yet to be described. Here, we microinjected and expressed various snRNA deletion mutants as well as chimeric 7SK, Alu or bacterial SRP non-coding RNAs and provide evidence that Sm and SMN binding sites are necessary and sufficient for CB localization of snRNAs. We further show that Sm proteins, and specifically their GR-rich domains, are important for accumulating snRNPs in CBs. Accordingly, core snRNPs containing the Sm proteins, but not naked snRNAs, restore the formation of CBs after their depletion. Finally, we show that immature but not fully assembled snRNPs are able to induce CB formation and that microinjection of an excess of U2 snRNP-specific proteins, which promotes U2 snRNP maturation, chases U2 snRNA from CBs. We propose that the accessibility of the Sm ring represents the molecular basis for the quality control of the final maturation of snRNPs and the sequestration of immature particles in CBs.
Figures

Similar articles
-
A role for Cajal bodies in the final steps of U2 snRNP biogenesis.J Cell Sci. 2004 Sep 1;117(Pt 19):4423-33. doi: 10.1242/jcs.01308. Epub 2004 Aug 17. J Cell Sci. 2004. PMID: 15316075
-
The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze.Chromosoma. 2006 Oct;115(5):343-54. doi: 10.1007/s00412-006-0056-6. Epub 2006 Mar 31. Chromosoma. 2006. PMID: 16575476 Review.
-
RNA-mediated interaction of Cajal bodies and U2 snRNA genes.J Cell Biol. 2001 Aug 6;154(3):499-509. doi: 10.1083/jcb.200105084. J Cell Biol. 2001. PMID: 11489914 Free PMC article.
-
Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies.Mol Biol Cell. 2006 Jul;17(7):3221-31. doi: 10.1091/mbc.e06-03-0247. Epub 2006 May 10. Mol Biol Cell. 2006. PMID: 16687569 Free PMC article.
-
Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos.RNA Biol. 2017 Jun 3;14(6):752-760. doi: 10.1080/15476286.2016.1255397. Epub 2016 Nov 18. RNA Biol. 2017. PMID: 27858508 Free PMC article. Review.
Cited by
-
Biology of the mRNA Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities.Int J Mol Sci. 2021 May 12;22(10):5110. doi: 10.3390/ijms22105110. Int J Mol Sci. 2021. PMID: 34065983 Free PMC article. Review.
-
Are Supermeres a Distinct Nanoparticle?J Extracell Biol. 2022 Jun;1(6):e44. doi: 10.1002/jex2.44. Epub 2022 Jun 3. J Extracell Biol. 2022. PMID: 36311879 Free PMC article.
-
Acetylation-dependent regulation of core spliceosome modulates hepatocellular carcinoma cassette exons and sensitivity to PARP inhibitors.Nat Commun. 2024 Jun 18;15(1):5209. doi: 10.1038/s41467-024-49573-7. Nat Commun. 2024. PMID: 38890388 Free PMC article.
-
The SMN complex drives structural changes in human snRNAs to enable snRNP assembly.Nat Commun. 2023 Oct 18;14(1):6580. doi: 10.1038/s41467-023-42324-0. Nat Commun. 2023. PMID: 37852981 Free PMC article.
-
The 3' Pol II pausing at replication-dependent histone genes is regulated by Mediator through Cajal bodies' association with histone locus bodies.Nat Commun. 2022 May 25;13(1):2905. doi: 10.1038/s41467-022-30632-w. Nat Commun. 2022. PMID: 35614107 Free PMC article.
References
-
- Eggert C., Chari A., Laggerbauer B., Fischer U.. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006; 12:113–121. - PubMed
-
- Matera A.G., Shpargel K.B.. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 2006; 18:317–324. - PubMed
-
- Gruss O.J., Meduri R., Schilling M., Fischer U.. UsnRNP biogenesis: mechanisms and regulation. Chromosoma. 2017; 126:577–593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

