Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression
- PMID: 29386015
- PMCID: PMC5793418
- DOI: 10.1186/s12943-018-0771-7
Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression
Abstract
Background: Circ-ITCH is a circRNA generated from several exons of itchy E3 ubiquitin protein ligase (ITCH) and tumor suppressor served as a sponge for certain miRNAs targeting their parental transcripts of ITCH. However, the role of circ-ITCH in bladder cancer (BCa) was not reported. In the present study, we investigated the role of circ-ITCH in BCa.
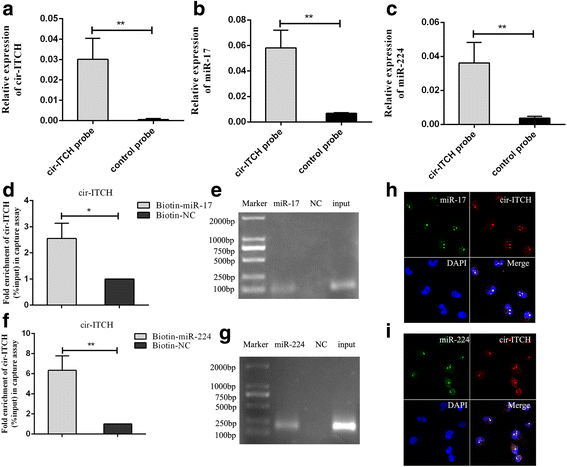
Methods: Quantitative real-time PCR was used to detect the expression of circ-ITCH and survival analysis was adopted to explore the association between circ-ITCH expression and the prognosis of BCa. BCa cells were stably transfected with lentivirus approach and cell proliferation, migration, invasion, cell cycle and cell apoptosis, as well as tumorigenesis in nude mice were performed to assess the effect of circ-ITCH in BCa. Biotin-coupled probe pull down assay, Biotin-coupled miRNA capture, Fluorescence in situ hybridization and Luciferase reporter assay were conducted to confirm the relationship between the circ-ITCH and the microRNA.
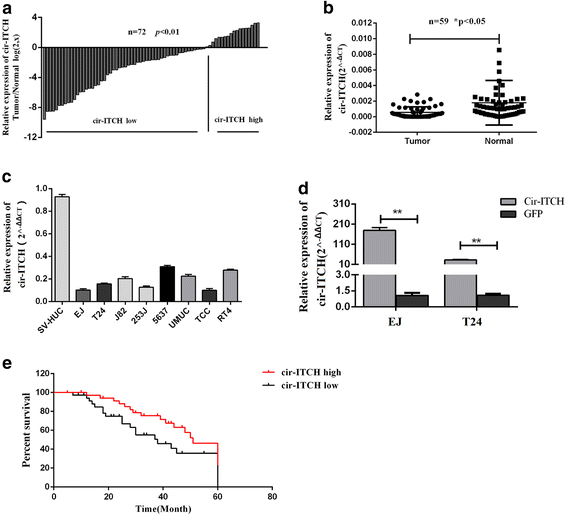
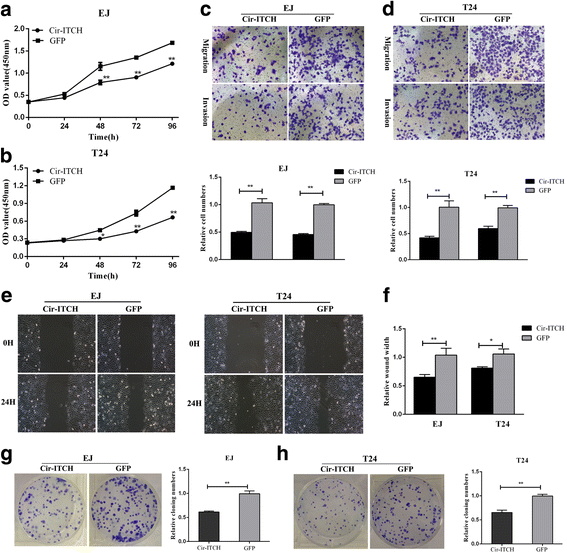
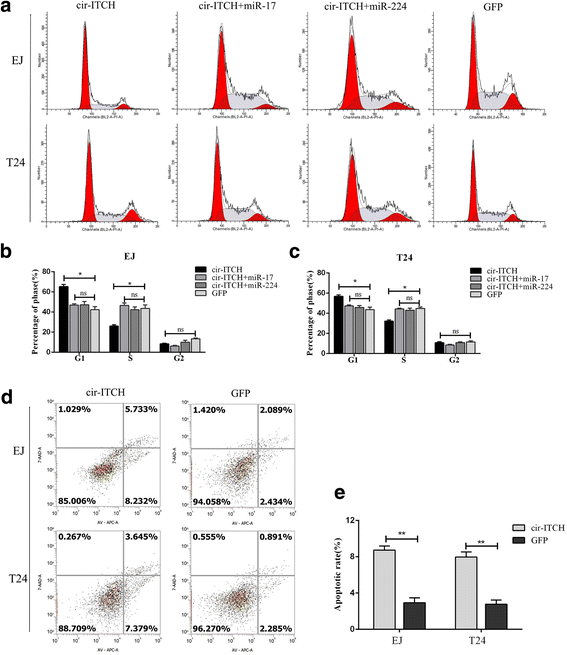
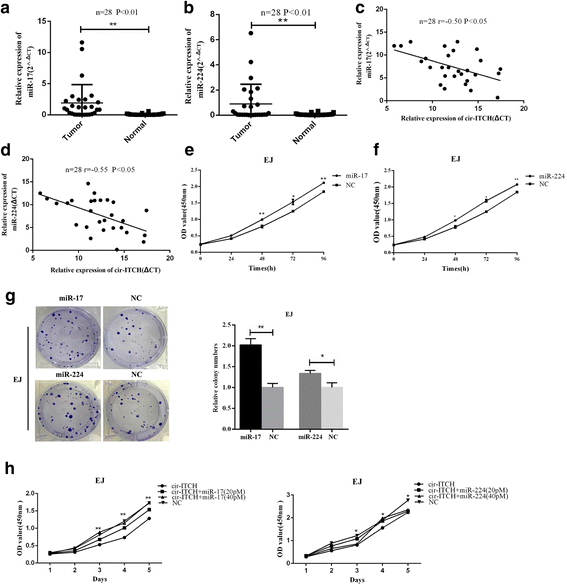
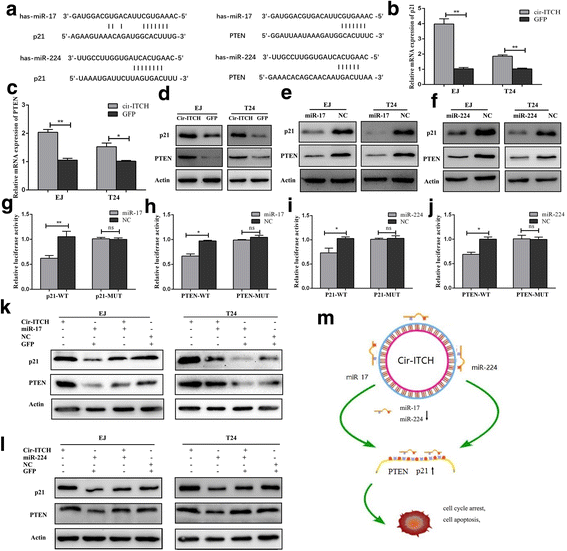
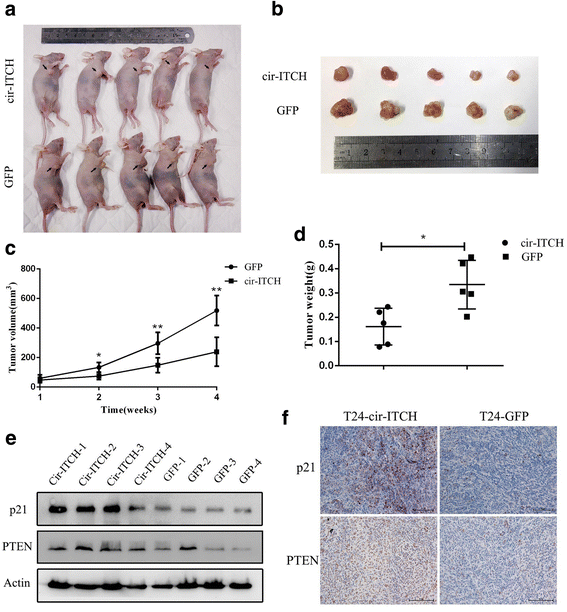
Results: In the present study, we found that circ-ITCH, is down-regulated in BCa tissues and cell lines. BCa patients with low circ-ITCH expression had shortened survival. Enforced- expression of circ-ITCH inhibited cells proliferation, migration, invasion and metastasis both in vitro and in vivo. Mechanistically, we demonstrated that circ-ITCH up-regulates the expression of miR-17 and miR-224 target gene p21 and PTEN through 'sponging' miR-17 and miR-224, which suppressed the aggressive biological behaviors of BCa.
Conclusions: circ-ITCH acts as a tumor suppressor by a novel circ-ITCH/miR-17, miR-224/p21, PTEN axis, which may provide a potential biomarker and therapeutic target for the management of BCa.
Keywords: Bladder cancer; PTEN; circ-ITCH; miR-17; miR-224; p21.
Conflict of interest statement
Ethics approval and consent to participate
The present study was approved by the Hospital’s Protection of Human Subjects Committee.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence.Mol Cancer. 2019 Sep 3;18(1):133. doi: 10.1186/s12943-019-1060-9. Mol Cancer. 2019. PMID: 31481066 Free PMC article.
-
Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression.J Exp Clin Cancer Res. 2019 Apr 18;38(1):169. doi: 10.1186/s13046-019-1136-9. J Exp Clin Cancer Res. 2019. PMID: 30999937 Free PMC article.
-
Circular RNA ITCH attenuates the progression of nasopharyngeal carcinoma by inducing PTEN upregulation via miR-214.J Gene Med. 2022 Jan;24(1):e3391. doi: 10.1002/jgm.3391. Epub 2021 Nov 11. J Gene Med. 2022. PMID: 34612550
-
Circular RNA ITCH: A novel tumor suppressor in multiple cancers.Life Sci. 2020 Aug 1;254:117176. doi: 10.1016/j.lfs.2019.117176. Epub 2019 Dec 13. Life Sci. 2020. PMID: 31843532 Review.
-
Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer.Mol Cancer. 2021 Jan 4;20(1):4. doi: 10.1186/s12943-020-01300-8. Mol Cancer. 2021. PMID: 33397425 Free PMC article. Review.
Cited by
-
Has_circ_0002360 promotes the progression of lung adenocarcinoma by activating miR-762 and regulating PODXL expression.Transl Cancer Res. 2024 Aug 31;13(8):4172-4186. doi: 10.21037/tcr-24-279. Epub 2024 Aug 27. Transl Cancer Res. 2024. PMID: 39262484 Free PMC article.
-
hsa_circ_0001018 promotes papillary thyroid cancer by facilitating cell survival, invasion, G1/S cell cycle progression, and repressing cell apoptosis via crosstalk with miR-338-3p and SOX4.Mol Ther Nucleic Acids. 2021 Feb 24;24:591-609. doi: 10.1016/j.omtn.2021.02.023. eCollection 2021 Jun 4. Mol Ther Nucleic Acids. 2021. PMID: 33898108 Free PMC article.
-
Extracellular vesicle-mediated delivery of miR-766-3p from bone marrow stromal cells as a therapeutic strategy against colorectal cancer.Cancer Cell Int. 2024 Oct 1;24(1):330. doi: 10.1186/s12935-024-03493-0. Cancer Cell Int. 2024. PMID: 39354491 Free PMC article.
-
Circular RNA circGRAMD1B inhibits gastric cancer progression by sponging miR-130a-3p and regulating PTEN and p21 expression.Aging (Albany NY). 2019 Nov 13;11(21):9689-9708. doi: 10.18632/aging.102414. Epub 2019 Nov 13. Aging (Albany NY). 2019. PMID: 31719211 Free PMC article.
-
Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis.Cancer Med. 2020 Jan;9(2):700-710. doi: 10.1002/cam4.2650. Epub 2019 Nov 23. Cancer Med. 2020. PMID: 31758655 Free PMC article.
References
-
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- JX10231801/the Priority Academic Program Development of Jiangsu Higher Education Institutions/International
- BE2016791/the Provincial Initiative Program for Excellency Disciplines of Jiangsu Province/International
- 81272832/the National Natural Science Foundation of China/International
- 81602235/the National Natural Science Foundation of China/International
- 81772711/the National Natural Science Foundation of China/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

