Cell-specific responses to the cytokine TGFβ are determined by variability in protein levels
- PMID: 29371237
- PMCID: PMC5787704
- DOI: 10.15252/msb.20177733
Cell-specific responses to the cytokine TGFβ are determined by variability in protein levels
Abstract
The cytokine TGFβ provides important information during embryonic development, adult tissue homeostasis, and regeneration. Alterations in the cellular response to TGFβ are involved in severe human diseases. To understand how cells encode the extracellular input and transmit its information to elicit appropriate responses, we acquired quantitative time-resolved measurements of pathway activation at the single-cell level. We established dynamic time warping to quantitatively compare signaling dynamics of thousands of individual cells and described heterogeneous single-cell responses by mathematical modeling. Our combined experimental and theoretical study revealed that the response to a given dose of TGFβ is determined cell specifically by the levels of defined signaling proteins. This heterogeneity in signaling protein expression leads to decomposition of cells into classes with qualitatively distinct signaling dynamics and phenotypic outcome. Negative feedback regulators promote heterogeneous signaling, as a SMAD7 knock-out specifically affected the signal duration in a subpopulation of cells. Taken together, we propose a quantitative framework that allows predicting and testing sources of cellular signaling heterogeneity.
Keywords: TGFβ‐SMAD signaling; cellular heterogeneity; mathematical modeling; signaling dynamics; single‐cell analysis.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

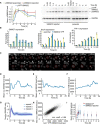
- A
Fluorescent reporter system to measure SMAD signaling dynamics in individual cells. SMAD2 was fused to the yellow fluorescent protein mVenus (YFP) under the control of the human ubiquitin C promoter (UbCp) with the selection marker G418 (Geneticin). As a nuclear marker, histone 2B (H2B) was fused to the cyan fluorescent protein mCerulean (CFP) under the control of UbCp with the selection marker hygromycin.
- B
Western blot analysis of endogenous and YFP‐tagged SMAD2 in a stable clonal reporter cell line and the corresponding parental cell line. Cells were stimulated with 100 pM TGFβ1 and analyzed after 3 h. GAPDH was used as a loading control.
- C
Western blot analysis of SMAD2 activation in SMAD2‐YFP reporter and parental MCF10A cells. Cells were stimulated with 100 pM TGFβ1, and SMAD2 phosphorylation was analyzed at indicated time points. GAPDH was used as a loading control.
- D, E
Live‐cell time‐lapse microscopy images of MCF10A cells expressing SMAD2‐YFP following treatment with 100 pM TGFβ1 (D). White circles indicate the segmented nucleus, and the estimated cytoplasmic area is represented by red annuli. The indicated cell was tracked over 24 h and the corresponding nuclear‐to‐cytoplasmic (nuc/cyt) SMAD2‐YFP ratio plotted over time (E).
- F
Time‐resolved analysis of the SMAD2 nuclear to cytoplasmic localization for eight individual cells (thin lines) compared to the median nuc/cyt SMAD2 ratio of the entire population (thick line) upon stimulation with 100 pM TGFβ1. See Appendix Table S1 for number of cells analyzed.
- G
Median nuc/cyt SMAD2 ratio for reporter cells stimulated with 100 pM TGFβ1 and treated with TGBβRI kinase inhibitor (SB431542) at indicated time points. At all time points, SMAD2 nuclear translocation was dependent on TGFβ receptor activity. See Appendix Table S1 for number of cells analyzed.
- A
Western blot analysis of SMAD2 activation in SMAD2‐YFP reporter and parental MCF10A cells. Cells were stimulated with 100 pM TGFβ1 and SMAD2 phosphorylation was analyzed at indicated time points. GAPDH was used as a loading control. Independent experiments were quantified and normalized to maximum values. Error bars indicate standard deviation of biological repeats (n = 4). Note that phosphorylated YFP‐SMAD2 was at background levels at 0.25 h presumably due to lower expression levels.
- B
Expression of SMAD target genes in parental and SMAD2 reporter cell lines. Expression kinetics of the SMAD target genes SMAD7, SnoN, and PAI‐1 upon 100 pM TGFβ stimulation were measured by qPCR in the indicated cell lines. β‐Actin was used as an internal control. Error bars indicate standard deviation of technical triplicates.
- C
Live‐cell time‐lapse microscopy images of H2B‐CFP expression in MCF10A cells following treatment with 100 pM TGFβ. The same detail as in Fig 1D is shown. White circle indicates the segmented nucleus, and the estimated cytoplasmic area is represented by red annuli.
- D–F
The cell indicated in Fig 1D was tracked over 24 h. The mean nuclear (D) and cytoplasmic (E) fluorescence intensity of SMAD2‐YFP as well as the nuclear fluorescence intensity of H2B‐CFP (F) were measured upon 100 pM TGFβ stimulation.
- G
Reproducibility of SMAD2 translocation measurements. Median SMAD2‐YFP ratios (solid lines) of cells plated in three independent glass bottom plates stimulated with 100 pM TGFβ at the same day and tracked over 24 h (biological triplicates). Shaded areas indicate 25th and 75th percentiles. See Appendix Table S1 for number of cells analyzed.
- H
Correlation between endogenously expressed SMAD2 and transgenic YFP‐SMAD2. In the same individual SMAD2 reporter cells treated with 100 pM TGFβ for 1.5 h, nuclear endogenous SMAD2 was measured by immunofluorescence and compared to the nuclear fluorescence intensity from YFP‐SMAD2. Both measures were highly correlated (Pearson's correlation, n = 7,300).
- I
Comparison of endogenous SMAD2 activation and SMAD2‐YFP translocation. The nuc/cyt ratio of SMAD2‐YFP upon 100 pM TGFβ stimulation was measured in reporter cells by time‐lapse microscopy at the indicated time points (blue); phosphorylation of endogenous SMAD2 was measured in parental MCF10A cells by immunofluorescence (IF, red) under the same conditions. Data were normalized by minimum subtraction and division through the overall maximum. White and black dots indicate medians; boxes include data between the 25th and 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; colored dots represent outliers. See Appendix Table S1 for number of cells analyzed.
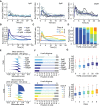
Time‐resolved analysis of SMAD2 nuclear to cytoplasmic localization for varying stimulus levels. Nuc/cyt SMAD2 ratios for eight individual cells (thin lines) as well as the population median (thick line) are shown. See Appendix Table S1 for number of cells analyzed.
Median nuc/cyt SMAD2 ratio of cells stimulated with varying concentrations of TGFβ1 over 24 h. See Appendix Table S1 for number of cells analyzed.
Individual cells were clustered into six signaling classes according to their time‐resolved nuc/cyt SMAD2 ratio using dynamic time warping (DTW). Each line represents the median over all cells of the indicated cluster. Cells stimulated with varying TGFβ1 concentrations as indicated in (B) were included in the analysis.
Distributions of signaling classes depending on TGFβ dose.
Silhouette plots of cells sorted according to TGFβ concentration (upper panel) or signaling classes (lower panel). Plots provide a graphical representation of how well the nuc/cyt SMAD2 ratios of each cell correspond to trajectories of other cells in its own group according to the cDTW measure. Positive silhouette scores indicate that SMAD2 responses are more similar to the own group, while negative scores signify that the corresponding trajectory is closer to any of the other groups. In general, signaling classes provide better separation than sorting according to stimulus levels.
Cell proliferation shown as number of cell divisions per cell within 24 h after a TGFβ stimulus. Cells were sorted according to TGFβ concentrations (upper panel) or signaling classes (lower panel).
Motility of each cell as summed distance covered between 20 and 24 h after stimulation with TGFβ (in pixel). Cells were sorted according to TGFβ concentrations (upper panel) or signaling classes (lower panel). White lines indicate median; boxes include data between the 25th and 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; crosses represent outliers. See Appendix Table S1 for number of cells analyzed.
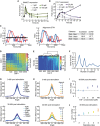
- A, B
Dose‐dependency of SMAD target gene expression. Parental MCF10A cells were stimulated with varying concentrations of TGFβ and SMAD7 (A) and PAI‐1 (B) expression was measured by qPCR at indicated time points. Error bars indicate standard deviation of technical triplicates.
- C
Illustrative comparison of Euclidian distance and dynamic time warping (DTW). In the left panel, three single‐cell trajectories (red, blue, and black) with similar Euclidian distances are shown (see table). DTW performs a non‐linear alignment in time (middle panel) that compensates the temporal shift of the peaks in the red and blue trajectory (cyan lines). This leads to a lower DTW distance score compared to the DTW distance between the red and black or blue and black trajectories, which remain almost unchanged (see table).
- D
Dissimilarity matrix calculated pair‐wise by cDTW of single‐cell trajectories treated with varying TGFβ doses. Strength of the TGFβ stimulation increases from top left to bottom right.
- E
Heat map of single‐cell time courses sorted according to hierarchical clustering. The corresponding dendrogram is shown on top.
- F
Optimal number of clusters. For different cluster numbers, jump size is calculated using sum of square errors of cDTW scores as a measure of intra‐cluster dispersion. Jump size reaches maxima at three and six clusters, indicating that these are good choices for cluster number.
- G–J
Direction‐resolved analysis of cell motility in TGFβ‐stimulated cells. The angle and distance of each cell movement were determined and averaged for 0–30 h (upper panel) and 30–60 h (lower panel) after stimulation with varying concentrations of TGFβ (see Appendix II.F for details). Cells are grouped according to stimulation levels (G) or signaling classes (H). Changes in cell motility are more pronounced at later time points after stimulation. Unidirectional movements (angle = 0) of TGFβ‐stimulated cells 30–60 h after treatment were normalized by the mean movement of unstimulated cells in the same time period and analyzed according to stimulus level (I) or signaling classes (J). Changes in cell motility are express as median fold change; error bars indicate 95% confidence intervals from permutation testing. Signaling dynamics allow better stratification of cellular outcomes compared to stimulus levels. See Appendix Table S1 for number of cells analyzed.
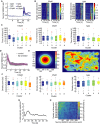
Time‐resolved analysis of SMAD2 nuclear/cytoplasmic localization before and after stimulation with varying concentrations of TGFβ. Cells were imaged for 24 h before and 12 h after stimulation. Median nuc/cyt SMAD2 ratios for indicated stimulation levels are shown. See Appendix Table S1 for number of cells analyzed.
Heat map of SMAD2 translocation in individual cells imaged for 24 h before stimulation with 25 pM or 5 pM TGFβ (similar to Fig 3A). Each horizontal line represents a single cell. Cells were sorted either by the time of the last division before stimulation (left) or by the amplitude of their response (right). Obviously, the time of last division and signaling responses are not correlated. See Appendix Table S1 for number of cells analyzed.
Time of last cell division before stimulus for each signaling class after stimulation with the indicated concentrations of TGFβ. Single‐cell time courses were mapped onto the previously observed signaling classes (Fig 2C; see Appendix II.H). Distributions are overlapping; no significant trend in cell division time is observable. White lines indicate median; boxes include data between the 25th and the 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; crosses represent outliers. See Appendix Table S1 for number of cells analyzed.
SMAD2 response in G2 arrested cells. Cells were arrested in G2 using the CDK1 inhibitor RO3306, stimulated with 100 pM TGFβ and followed for 24 h by live‐cell imaging. No difference to control cells treated with vehicle could be observed. Solid lines indicate median nuc/cyt SMAD2 ratios, shaded areas 25th to 75th percentiles. See Appendix Table S1 for number of cells analyzed.
Measuring local cell density by live‐cell imaging. Local cell density is measured in a 200‐px radius around each cell for each time point by applying a bell‐shaped kernel to obtain a weighted sum of all neighboring cells (left). The resulting density scores are demonstrated using a randomly chosen time point. Red circles indicate the centroid of cells identified. Cells highlighted by blue circles were successfully tracked for the time of the experiment. Warmer colors indicate higher density scores.
Cell density before stimulation shown for each signaling class observed in response to stimulation with the indicated concentrations of TGFβ. Distributions of density scores are overlapping; no significant trend in cell density is observable. White lines indicate median; boxes include data between the 25th and the 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; crosses represent outliers. See Appendix Table S1 for number of cells analyzed.
Quantifying the contribution of cell cycle state to heterogeneity in SMAD signaling. Mutual information between time of last cell division and nuc/cyt SMAD ratio after stimulation with varying doses of TGFβ was determined for each time point and normalized by the sum of entropies to calculate the fraction of heterogeneity in SMAD signaling that can be explained by cell cycle state (Appendix II.E).
Quantifying the contribution of cell density to heterogeneity in SMAD signaling. Mutual information between cell density scores and nuc/cyt SMAD ratio after stimulation with varying doses of TGFβ was determined pair‐wise for all combinations of time point and normalized by the sum of entropies to calculate the fraction of heterogeneity in SMAD signaling that can be explained by cell density at any time point. The corresponding heat map demonstrates that cell density provides only a minor contribution to heterogeneity.
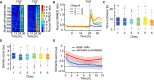
Heat map of SMAD2 translocation in individual cells over time. Cells were imaged for 24 h before stimulation with 100 pM TGFβ1. Each horizontal line represents a single cell, and the nuc/cyt ratio is shown as indicated in the legend. Time of cell division is indicated by white marks. Cells were sorted either by the time of the last division before stimulation (left) or by the amplitude of their response (right). Cell cycle and response are not correlated. See Appendix Table S1 for number of cells analyzed.
Mapping of SMAD2 translocation dynamics in individual cells to previously identified signaling classes (compare Fig 2C). Cells were imaged for 24 h before stimulation with varying TGFβ1 concentrations (Fig EV3A). For each trajectory, the most similar signaling class was determined using Euclidian distance to the median dynamics of the previously defined clusters (Fig 2C) as a similarity measure. Median nuc/cyt SMAD2 ratios for resulting mapped subpopulations are shown. See Appendix Table S1 for number of cells analyzed.
Time of last cell division before stimulus for each signaling class (defined in B). Distributions are overlapping; no significant trend in cell division time is observable. White lines indicate median; boxes include data between the 25th and 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; crosses represent outliers. See Appendix Table S1 for number of cells analyzed.
Cell density before stimulus for each signaling class (defined in B). Density scores represent a weighted sum of all neighboring cells within 640 μm distance. Distributions are overlapping; no significant trend in cell density is observable. White lines indicate median; boxes include data between the 25th and 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; crosses represent outliers. See Appendix Table S1 for number of cells analyzed.
Analysis of SMAD2 translocation dynamics in sister cells. SMAD2 translocation dynamics in sister cells after division and unrelated cell pairs with the same nuc/cyt SMAD2 ratio were compared using cDTW. Resulting similarity scores were aligned in time and compared to those from randomly selected cell pairs. Effect size (solid lines) and 95% confidence intervals (shaded areas) were estimated by bootstrapping. The analysis shows that recently divided cells are more similar than control cell pairs and remain correlated over time, indicating that heterogeneity arises from differences in cellular state. See Appendix Table S1 for number of cells analyzed.

- A
Outline of a tiered approach to model heterogeneous signaling in single cells (see text for details).
- B
Topology of TGFβ pathway model. The oval shapes represent free receptors (blue), ligand (yellow), and ligand–receptor complex (gray). Extension “‐e” signifies endosomal species. Rectangles represent SMAD2 (blue), SMAD4 (green), and generic feedback regulator (yellow). Extensions “p” indicate phosphorylated and “n” nuclear species. Production and degradation are shown by phi symbols. State transitions and intercompartmental shuttling are indicated with arrows, enzyme catalysis with circle headed bars, and feedback inhibition with blunt headed bars.
- C
Calibration of population‐average model by fitting to median SMAD2 translocation dynamics of cells stimulated with different TGFβ concentrations. Experimental data points correspond to Fig 2B. Model fits to other datasets are shown in Fig EV4 (see also Appendix Table S4); parameter values are provided in Appendix Table S5 and Table EV1.
- D
Medium TGFβ degradation over time. Blue line shows the ligand concentration after an initial stimulus with 25 pM TGFβ1 as predicted by the best‐fit mathematical model. Shaded area represents the range of predictions from 30 fits with similar goodness of fit obtained from local multistart optimization (see Appendix III.D). Black stars indicate corresponding experimental measurements. Error bars represent standard deviation from three replicates.
- E–G
Time‐dependent restimulation of the TGFβ pathway at varying input levels. Measured median nuc/cyt SMAD2 ratios (*) and model predictions (−) are shown. Solid lines represent the best‐fit model and shaded areas the range of predictions from 30 independent fits (see D). Dashed vertical lines indicate time of second stimulus, which replenishes the extracellular ligand pool to its initial concentration. (E) 5 pM TGFβ1 was applied at 0 h and 3 h. (F) 5 pM TGFβ was applied at 0 h and 8 h. (G) 100 pM TGFβ1 was applied at 0 h and 8 h. See Appendix Table S1 for number of cells analyzed.
- H
Effect of the global transcriptional inhibitor DRB on SMAD signaling. Cells were stimulated with 100 pM TGFβ1 in the presence or absence of DRB. Measured median nuc/cyt SMAD2 ratios (*) and model predictions (−) are shown. Solid line represents the best‐fit model and shaded area the range of predictions from 30 independent fits (see D). See Appendix Table S1 for number of cells analyzed.
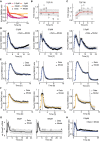
- A
Model calibration with median SMAD4 translocation dynamics of cells stimulated with different TGFβ concentrations. Solid lines represent model fit, filled circles measured data. Experimental data points correspond to Appendix Fig S2J.
- B, C
Dynamics of total cellular TGFβ receptor protein levels upon stimulation with 100 pM TGFβ. Dynamics of TGFβR1 (B) and TGFβR2 (C) are shown as fold change relative to unstimulated cells. Solid red lines represent model fit and circles data measured by Western blot analysis. For each observation point, data spread from triplicates is shown. Gray area indicates measurement noise estimated by the fitting algorithm.
- D
Restimulation with 2.5 pM TGFβ. Stimulation was performed once at the beginning of the experiment (left), at the beginning and at 3 h (middle), or at the beginning and at 6 h (right) to replenish the ligand pool to its initial concentration. Solid lines represent model fits, and circles measured data points. Gray area indicates measurement noise estimated by the fitting algorithm.
- E, F
Model calibration with median SMAD2 and SMAD4 translocation dynamics upon receptor inhibition. Cells were stimulated with 100 pM TGFβ, TGFβRI activity was inhibited using the small‐molecule receptor inhibitor SB431542 at the indicated time points (dashed vertical lines). Nuc/cyt SMAD2 (E) and SMAD4 (F) ratios were considered. Solid lines represent model fits, and circles or asterisks measured data. Experimental data points correspond to Fig 1G and Appendix Fig S2K. Gray area indicates measurement noise estimated by the fitting algorithm.
- G
Smad7 mRNA induction upon 25 pM and 100 pM TGFb stimulation. Solid lines represent model fits, and circles measured fold changes relative to unstimulated cells. Experimental data points correspond to Fig EV2A. Gray area indicates measurement noise estimated by the fitting algorithm.
- H
Model prediction of restimulation with 100 pM TGFβ at 3 h (dashed vertical line). Solid line represents model prediction, and asterisks (*) measured data.
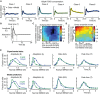
- A
The model of TGFβ signaling was fitted to six signaling classes observed upon stimulation with 100 pM TGFβ1. Median nuc/cyt SMAD2 ratios (circles) and model fits (solid lines) are shown.
- B
Features of SMAD2 translocation dynamics. We considered the amplitude (E) and timing (T) of the first peak of nuclear translocation as well as the amplitude at 300 min (L) as a measure for the signaling activity upon adaptation of the pathway.
- C, D
Model performance at varying noise levels. Heterogeneous signaling in response to a 100 pM TGFβ1 stimulus was simulated by signaling class‐based modeling (C) or a direct ensemble modeling (D) (see main text). Noise in protein expression is modeled as a combination of correlated and uncorrelated noise (see Appendix IV.B). The differences among single‐cell signaling features between model and data are calculated as sum of squared errors and normalized to the maximal deviation observed (color bar). For each combination of correlated and uncorrelated noise, 10,000 cells were simulated.
- E
Measured and predicted distributions of signaling features for two TGFβ stimuli (2.5 and 100 pM). A population of artificial cells was assembled according to signaling class distributions observed upon stimulation with 100 pM TGFβ1 using optimal noise contributions (see panel C). Signaling features were extracted from simulations at different TGFβ concentrations.

Unimodal distribution of protein concentrations in artificial cell populations. Distributions of basal concentrations for TGFβR1, SMAD2, and SMAD4 are shown for a population of artificial cells assembled according to the observed proportion of signaling classes at 100 pM TGFβ using calibrated noise levels.
Silhouette plots of artificial cells sorted according to TGFβ concentration (left panel) or mapped to experimentally observed signaling classes (right panel). Plots provide a graphical representation of how well the nuc/cyt SMAD2 ratios of each simulated cell corresponds to trajectories of other simulated cells in its own group. Positive silhouette scores indicate that SMAD2 responses are more similar to the own group, while negative scores signify that the corresponding trajectory is closer to any of the other groups. In general, signaling classes provide better separation than sorting according to stimulus levels.
Transition between signaling classes depending on feedback strength. The response of a reassembled population of artificial cells to 5 and 25 pM TGFβ was simulated with reduced feedback expression as indicated and mapped to previously observed signaling classes (see Appendix II.H). Black lines and their thickness indicate the direction and extent of transitions between signaling classes. Transitions with a probability below 1% are excluded for better visualization.
Robustness of model predictions concerning SMAD7 knock‐out effect on distribution of signaling classes at 100 pM (top) and 5 pM (bottom) TGFβ, respectively. The simulated fraction of cells in each signaling class in wt and 30% feedback depleted cells is shown for 30 independent fits with similar goodness of fit obtained from local multistart optimization (see Appendix IV.D). The corresponding best‐fit results are shown in Fig 6A and G. Artificial cell populations were generated by adding the same protein concentration noise as in the best‐fit model (Fig 5C). White lines indicate median; boxes include data between the 25th and 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range; crosses represent outliers.
Sequence of SMAD7 knock‐out alleles. The indicated sequence (red) in the second exon of the SMAD7 gene was targeted by Cas9 in combination with a specific sgRNA. This led to deletions of 22 nt and 7 nt in the targeted alleles, causing frameshifts and non‐sense mutations in the SMAD7 gene.
Expression of SMAD7 mRNA in parental and knock‐out cells. Basal SMAD7 mRNA levels were determined by qPCR in the indicated cell lines. SMAD7 mRNA containing a prematures stop codon is degraded in knock‐out cells due to non‐sense mediated decay. β‐Actin was used as an internal control. Error bars indicate standard deviation of technical triplicates.
Time‐resolved analysis of SMAD2 nuclear/cytoplasmic translocation in the absence of SMAD7. Median nuc/cyt SMAD2 ratio of cells stimulated with varying concentrations of TGFβ over 24 h are shown for parental (left) and SMAD7 knock‐out cells. See Appendix Table S1 for number of cells analyzed.

Predicted distributions of signaling classes depending on TGFβ dose. Simulations were performed as described for Fig 5E. The simulated time courses were mapped onto the original clusters dynamics (Fig 2C) as described in Appendix II.H.
Transition between signaling classes depending on stimulus strength. Same data as in (A). Black lines and their thickness indicate the direction and extent of transitions between signaling classes. Filled circle size indicates the proportion of artificial cells in the corresponding signaling class.
Transition between signaling classes depending on feedback strength. The response of a reassembled population of artificial cells to 100 pM TGFβ1 was simulated with reduced feedback expression as indicated (see Appendix IV.D) and mapped to previously observed signaling classes. Black lines and their thickness indicate the direction and extent of transitions between signaling classes. Transitions with a probability below 1% were excluded for better visualization.
Variation of model parameters across signaling classes. For 30 independent model fits to the experimentally observed signaling classes upon stimulation with 100 pM TGFβ1 (see Appendix), the variation of the indicated parameters between signaling classes was calculated as entropy. Lower entropies indicate more variation between signaling classes; uniform parameter distribution would lead to the maximal entropy of 2.6 bits. White lines indicate median; boxes include data between the 25th and 75th percentiles; whiskers extend to maximum values within 1.5× the interquartile range.
Distribution of signaling classes in parental and SMAD7 knock‐out cells. Cells were stimulated with indicated concentrations of TGFβ and measured SMAD2 translocation dynamics mapped to the previously observed signaling classes (Fig 2C).
Calibration of feedback level. Signaling class distributions at varying levels of feedback expression (C) were compared to experimentally observed distribution upon SMAD7 knock‐out (E). Minimal divergence between model and data was observed at 30% feedback expression.
Predicted distributions of signaling classes depending on TGFβ dose at 30% feedback expression. Simulations were mapped to the previously observed signaling classes (Fig 2C)
Similar articles
-
Modeling Cellular Signaling Variability Based on Single-Cell Data: The TGFβ-SMAD Signaling Pathway.Methods Mol Biol. 2023;2634:215-251. doi: 10.1007/978-1-0716-3008-2_10. Methods Mol Biol. 2023. PMID: 37074581 Review.
-
Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway.Cell. 2006 Jun 2;125(5):929-41. doi: 10.1016/j.cell.2006.03.045. Cell. 2006. PMID: 16751102
-
Smad signaling dynamics: insights from a parsimonious model.Sci Signal. 2008 Sep 9;1(36):pe41. doi: 10.1126/scisignal.136pe41. Sci Signal. 2008. PMID: 18780891
-
Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system.Proc Natl Acad Sci U S A. 2008 May 6;105(18):6608-13. doi: 10.1073/pnas.0710134105. Epub 2008 Apr 28. Proc Natl Acad Sci U S A. 2008. PMID: 18443295 Free PMC article.
-
Regulating the stability of TGFbeta receptors and Smads.Cell Res. 2009 Jan;19(1):21-35. doi: 10.1038/cr.2008.308. Cell Res. 2009. PMID: 19030025 Review.
Cited by
-
p53 dynamics in single cells are temperature-sensitive.Sci Rep. 2020 Jan 30;10(1):1481. doi: 10.1038/s41598-020-58267-1. Sci Rep. 2020. PMID: 32001771 Free PMC article.
-
State- and stimulus-specific dynamics of SMAD signaling determine fate decisions in individual cells.Proc Natl Acad Sci U S A. 2023 Mar 7;120(10):e2210891120. doi: 10.1073/pnas.2210891120. Epub 2023 Mar 1. Proc Natl Acad Sci U S A. 2023. PMID: 36857347 Free PMC article.
-
A systematic approach to decipher crosstalk in the p53 signaling pathway using single cell dynamics.PLoS Comput Biol. 2020 Jun 26;16(6):e1007901. doi: 10.1371/journal.pcbi.1007901. eCollection 2020 Jun. PLoS Comput Biol. 2020. PMID: 32589666 Free PMC article.
-
Modeling Cellular Signaling Variability Based on Single-Cell Data: The TGFβ-SMAD Signaling Pathway.Methods Mol Biol. 2023;2634:215-251. doi: 10.1007/978-1-0716-3008-2_10. Methods Mol Biol. 2023. PMID: 37074581 Review.
-
Transcriptional kinetic synergy: A complex landscape revealed by integrating modeling and synthetic biology.Cell Syst. 2023 Apr 19;14(4):324-339.e7. doi: 10.1016/j.cels.2023.02.003. Cell Syst. 2023. PMID: 37080164 Free PMC article.
References
-
- Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB (1994) An assay for transforming growth factor‐beta using cells transfected with a plasminogen activator inhibitor‐1 promoter‐luciferase construct. Anal Biochem 216: 276–284 - PubMed
-
- Adamson A, Boddington C, Downton P, Rowe W, Bagnall J, Lam C, Maya‐Mendoza A, Schmidt L, Harper CV, Spiller DG, Rand DA, Jackson DA, White MRH, Paszek P (2016) Signal transduction controls heterogeneous NF‐κB dynamics and target gene expression through cytokine‐specific refractory states. Nat Commun 7: 12057 - PMC - PubMed
-
- Bar‐Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

