Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions
- PMID: 29367353
- PMCID: PMC5839781
- DOI: 10.1083/jcb.201704184
Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions
Abstract
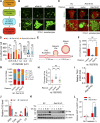
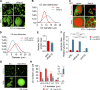
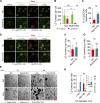
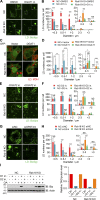
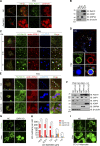
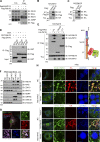
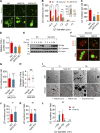
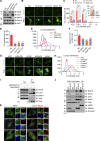
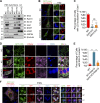
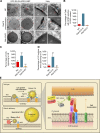
Lipid incorporation from endoplasmic reticulum (ER) to lipid droplet (LD) is important in controlling LD growth and intracellular lipid homeostasis. However, the molecular link mediating ER and LD cross talk remains elusive. Here, we identified Rab18 as an important Rab guanosine triphosphatase in controlling LD growth and maturation. Rab18 deficiency resulted in a drastically reduced number of mature LDs and decreased lipid storage, and was accompanied by increased ER stress. Rab3GAP1/2, the GEF of Rab18, promoted LD growth by activating and targeting Rab18 to LDs. LD-associated Rab18 bound specifically to the ER-associated NAG-RINT1-ZW10 (NRZ) tethering complex and their associated SNAREs (Syntaxin18, Use1, BNIP1), resulting in the recruitment of ER to LD and the formation of direct ER-LD contact. Cells with defects in the NRZ/SNARE complex function showed reduced LD growth and lipid storage. Overall, our data reveal that the Rab18-NRZ-SNARE complex is critical protein machinery for tethering ER-LD and establishing ER-LD contact to promote LD growth.
© 2018 Xu et al.
Figures
Similar articles
-
Characterization of the Role of Rab18 in Mediating LD-ER Contact and LD Growth.Methods Mol Biol. 2021;2293:229-241. doi: 10.1007/978-1-0716-1346-7_16. Methods Mol Biol. 2021. PMID: 34453721
-
Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells.Mol Biol Cell. 2018 Aug 15;29(17):2045-2054. doi: 10.1091/mbc.E18-05-0282. Epub 2018 Jun 27. Mol Biol Cell. 2018. PMID: 29949452 Free PMC article.
-
The ER-Localized Protein DFCP1 Modulates ER-Lipid Droplet Contact Formation.Cell Rep. 2019 Apr 9;27(2):343-358.e5. doi: 10.1016/j.celrep.2019.03.025. Cell Rep. 2019. PMID: 30970241
-
Come a little bit closer! Lipid droplet-ER contact sites are getting crowded.Biochim Biophys Acta Mol Cell Res. 2020 Feb;1867(2):118603. doi: 10.1016/j.bbamcr.2019.118603. Epub 2019 Nov 13. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 31733263 Review.
-
Born this way - Biogenesis of lipid droplets from specialized ER subdomains.Biochim Biophys Acta Mol Cell Biol Lipids. 2020 Jan;1865(1):158448. doi: 10.1016/j.bbalip.2019.04.008. Epub 2019 Apr 24. Biochim Biophys Acta Mol Cell Biol Lipids. 2020. PMID: 31028912 Review.
Cited by
-
Rab18 Drift in Lipid Droplet and Endoplasmic Reticulum Interactions of Adipocytes under Obesogenic Conditions.Int J Mol Sci. 2023 Dec 6;24(24):17177. doi: 10.3390/ijms242417177. Int J Mol Sci. 2023. PMID: 38139006 Free PMC article.
-
Protein Quality Control and Lipid Droplet Metabolism.Annu Rev Cell Dev Biol. 2020 Oct 6;36:115-139. doi: 10.1146/annurev-cellbio-031320-101827. Annu Rev Cell Dev Biol. 2020. PMID: 33021827 Free PMC article. Review.
-
Dynamics and functions of lipid droplets.Nat Rev Mol Cell Biol. 2019 Mar;20(3):137-155. doi: 10.1038/s41580-018-0085-z. Nat Rev Mol Cell Biol. 2019. PMID: 30523332 Free PMC article. Review.
-
ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets.J Cell Biol. 2020 Jan 6;219(1):e201905162. doi: 10.1083/jcb.201905162. J Cell Biol. 2020. PMID: 31653673 Free PMC article.
-
Relevance of Rab Proteins for the Life Cycle of Hepatitis C Virus.Front Cell Dev Biol. 2018 Dec 4;6:166. doi: 10.3389/fcell.2018.00166. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30564577 Free PMC article. Review.
References
-
- Ariotti N., Hall T.E., Rae J., Ferguson C., McMahon K.A., Martel N., Webb R.E., Webb R.I., Teasdale R.D., and Parton R.G.. 2015. Modular Detection of GFP-Labeled Proteins for Rapid Screening by Electron Microscopy in Cells and Organisms. Dev. Cell. 35:513–525. 10.1016/j.devcel.2015.10.016 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

