Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells
- PMID: 29335463
- PMCID: PMC5768742
- DOI: 10.1038/s41467-017-02526-9
Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells
Abstract
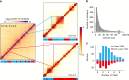
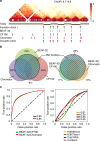
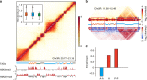
Topologically associating domains (TADs) are fundamental elements of the eukaryotic genomic structure. However, recent studies suggest that the insulating complexes, CTCF/cohesin, present at TAD borders in mammals are absent from those in Drosophila melanogaster, raising the possibility that border elements are not conserved among metazoans. Using in situ Hi-C with sub-kb resolution, here we show that the D. melanogaster genome is almost completely partitioned into >4000 TADs, nearly sevenfold more than previously identified. The overwhelming majority of these TADs are demarcated by the insulator complexes, BEAF-32/CP190, or BEAF-32/Chromator, indicating that these proteins may play an analogous role in flies as that of CTCF/cohesin in mammals. Moreover, extended regions previously thought to be unstructured are shown to consist of small contiguous TADs, a property also observed in mammals upon re-examination. Altogether, our work demonstrates that fundamental features associated with the higher-order folding of the genome are conserved from insects to mammals.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
High-resolution TADs reveal DNA sequences underlying genome organization in flies.Nat Commun. 2018 Jan 15;9(1):189. doi: 10.1038/s41467-017-02525-w. Nat Commun. 2018. PMID: 29335486 Free PMC article.
-
The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions.BMC Cell Biol. 2010 Dec 31;11:101. doi: 10.1186/1471-2121-11-101. BMC Cell Biol. 2010. PMID: 21194420 Free PMC article.
-
Quantitative differences in TAD border strength underly the TAD hierarchy in Drosophila chromosomes.J Cell Biochem. 2019 Mar;120(3):4494-4503. doi: 10.1002/jcb.27737. Epub 2018 Sep 27. J Cell Biochem. 2019. PMID: 30260021
-
Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion?: Alternating Chromatin States Provide a Basis for Domain Architecture in Drosophila.Bioessays. 2019 Sep;41(9):e1900048. doi: 10.1002/bies.201900048. Epub 2019 Jul 1. Bioessays. 2019. PMID: 31264253 Review.
-
Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review.
Cited by
-
Revisiting the organization of Polycomb-repressed domains: 3D chromatin models from Hi-C compared with super-resolution imaging.Nucleic Acids Res. 2020 Nov 18;48(20):11486-11494. doi: 10.1093/nar/gkaa932. Nucleic Acids Res. 2020. PMID: 33095877 Free PMC article.
-
Mechanisms of enhancer-promoter communication and chromosomal architecture in mammals and Drosophila.Front Genet. 2022 Dec 1;13:1081088. doi: 10.3389/fgene.2022.1081088. eCollection 2022. Front Genet. 2022. PMID: 36531247 Free PMC article. Review.
-
Cohesin controls intestinal stem cell identity by maintaining association of Escargot with target promoters.Elife. 2020 Feb 5;9:e48160. doi: 10.7554/eLife.48160. Elife. 2020. PMID: 32022682 Free PMC article.
-
Genomic organization of the autonomous regulatory domain of eyeless locus in Drosophila melanogaster.G3 (Bethesda). 2021 Dec 8;11(12):jkab338. doi: 10.1093/g3journal/jkab338. G3 (Bethesda). 2021. PMID: 34570231 Free PMC article.
-
3D or Not 3D: Shaping the Genome during Development.Cold Spring Harb Perspect Biol. 2022 May 27;14(5):a040188. doi: 10.1101/cshperspect.a040188. Cold Spring Harb Perspect Biol. 2022. PMID: 34312246 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

