ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma
- PMID: 29295999
- PMCID: PMC5750221
- DOI: 10.1038/s41467-017-02354-x
ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma
Erratum in
-
Publisher Correction: ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma.Nat Commun. 2018 Apr 6;9(1):1404. doi: 10.1038/s41467-018-03710-1. Nat Commun. 2018. PMID: 29626208 Free PMC article.
Abstract
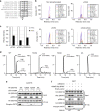
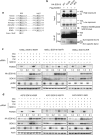
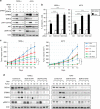
In human mutant BRAF melanoma cells, the stemness transcription factor FOXD3 is rapidly induced by inhibition of ERK1/2 signaling and mediates adaptive resistance to RAF inhibitors. However, the mechanism underlying ERK signaling control of FOXD3 expression remains unknown. Here we show that SOX10 is both necessary and sufficient for RAF inhibitor-induced expression of FOXD3 in mutant BRAF melanoma cells. SOX10 activates the transcription of FOXD3 by binding to a regulatory element in FOXD3 promoter. Phosphorylation of SOX10 by ERK inhibits its transcription activity toward multiple target genes by interfering with the sumoylation of SOX10 at K55, which is essential for its transcription activity. Finally, depletion of SOX10 sensitizes mutant BRAF melanoma cells to RAF inhibitors in vitro and in vivo. Thus, our work discovers a novel phosphorylation-dependent regulatory mechanism of SOX10 transcription activity and completes an ERK1/2/SOX10/FOXD3/ERBB3 axis that mediates adaptive resistance to RAF inhibitors in mutant BRAF melanoma cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Similar articles
-
LncRNA SAMMSON Mediates Adaptive Resistance to RAF Inhibition in BRAF-Mutant Melanoma Cells.Cancer Res. 2021 Jun 1;81(11):2918-2929. doi: 10.1158/0008-5472.CAN-20-3145. Epub 2021 Mar 18. Cancer Res. 2021. PMID: 34087780
-
The BRAF(V600E) inhibitor, PLX4032, increases type I collagen synthesis in melanoma cells.Matrix Biol. 2015 Oct;48:66-77. doi: 10.1016/j.matbio.2015.05.007. Epub 2015 May 16. Matrix Biol. 2015. PMID: 25989506 Free PMC article.
-
Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma.Nature. 2014 Apr 3;508(7494):118-22. doi: 10.1038/nature13121. Epub 2014 Mar 26. Nature. 2014. PMID: 24670642
-
ROCK1 is a potential combinatorial drug target for BRAF mutant melanoma.Mol Syst Biol. 2014 Dec 23;10(12):772. doi: 10.15252/msb.20145450. Mol Syst Biol. 2014. PMID: 25538140 Free PMC article.
-
Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells.Oncogene. 2012 May 10;31(19):2471-9. doi: 10.1038/onc.2011.424. Epub 2011 Sep 26. Oncogene. 2012. PMID: 21996740 Free PMC article.
Cited by
-
Clinical Correlation of Transcription Factor SOX3 in Cancer: Unveiling Its Role in Tumorigenesis.Genes (Basel). 2024 Jun 13;15(6):777. doi: 10.3390/genes15060777. Genes (Basel). 2024. PMID: 38927713 Free PMC article. Review.
-
Metabolism heterogeneity in melanoma fuels deactivation of immunotherapy: Predict before protect.Front Oncol. 2022 Dec 22;12:1046102. doi: 10.3389/fonc.2022.1046102. eCollection 2022. Front Oncol. 2022. PMID: 36620597 Free PMC article. Review.
-
Scoring model based on the signature of non-m6A-related neoantigen-coding lncRNAs assists in immune microenvironment analysis and TCR-neoantigen pair selection in gliomas.J Transl Med. 2022 Oct 29;20(1):494. doi: 10.1186/s12967-022-03713-z. J Transl Med. 2022. PMID: 36309750 Free PMC article.
-
CPT1A/2-Mediated FAO Enhancement-A Metabolic Target in Radioresistant Breast Cancer.Front Oncol. 2019 Nov 15;9:1201. doi: 10.3389/fonc.2019.01201. eCollection 2019. Front Oncol. 2019. PMID: 31803610 Free PMC article.
-
Signal pathways of melanoma and targeted therapy.Signal Transduct Target Ther. 2021 Dec 20;6(1):424. doi: 10.1038/s41392-021-00827-6. Signal Transduct Target Ther. 2021. PMID: 34924562 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

