CRISPR Screens Uncover Genes that Regulate Target Cell Sensitivity to the Morphogen Sonic Hedgehog
- PMID: 29290584
- PMCID: PMC5792066
- DOI: 10.1016/j.devcel.2017.12.003
CRISPR Screens Uncover Genes that Regulate Target Cell Sensitivity to the Morphogen Sonic Hedgehog
Erratum in
-
CRISPR Screens Uncover Genes that Regulate Target Cell Sensitivity to the Morphogen Sonic Hedgehog.Dev Cell. 2018 Jan 22;44(2):271. doi: 10.1016/j.devcel.2018.01.002. Dev Cell. 2018. PMID: 29401421 Free PMC article. No abstract available.
Abstract
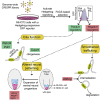
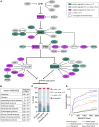
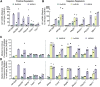
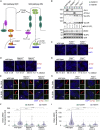
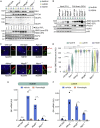
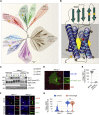
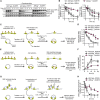
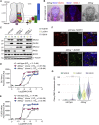
To uncover regulatory mechanisms in Hedgehog (Hh) signaling, we conducted genome-wide screens to identify positive and negative pathway components and validated top hits using multiple signaling and differentiation assays in two different cell types. Most positive regulators identified in our screens, including Rab34, Pdcl, and Tubd1, were involved in ciliary functions, confirming the central role for primary cilia in Hh signaling. Negative regulators identified included Megf8, Mgrn1, and an unannotated gene encoding a tetraspan protein we named Atthog. The function of these negative regulators converged on Smoothened (SMO), an oncoprotein that transduces the Hh signal across the membrane. In the absence of Atthog, SMO was stabilized at the cell surface and concentrated in the ciliary membrane, boosting cell sensitivity to the ligand Sonic Hedgehog (SHH) and consequently altering SHH-guided neural cell-fate decisions. Thus, we uncovered genes that modify the interpretation of morphogen signals by regulating protein-trafficking events in target cells.
Keywords: CRISPR screen; Hedgehog signaling; Smoothened; ciliopathy; congenital heart disease; heterotaxy; morphogen signaling; neural tube patterning; primary cilia; protein trafficking.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
A Membrane-Tethered Ubiquitination Pathway Regulates Hedgehog Signaling and Heart Development.Dev Cell. 2020 Nov 23;55(4):432-449.e12. doi: 10.1016/j.devcel.2020.08.012. Epub 2020 Sep 22. Dev Cell. 2020. PMID: 32966817 Free PMC article.
-
Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling.Mol Cell. 2017 Apr 6;66(1):154-162.e10. doi: 10.1016/j.molcel.2017.02.015. Epub 2017 Mar 23. Mol Cell. 2017. PMID: 28344083
-
G protein-coupled receptors control the sensitivity of cells to the morphogen Sonic Hedgehog.Sci Signal. 2018 Feb 6;11(516):eaao5749. doi: 10.1126/scisignal.aao5749. Sci Signal. 2018. PMID: 29438014 Free PMC article.
-
Mechanisms of Smoothened Regulation in Hedgehog Signaling.Cells. 2021 Aug 20;10(8):2138. doi: 10.3390/cells10082138. Cells. 2021. PMID: 34440907 Free PMC article. Review.
-
Signaling in the primary cilium through the lens of the Hedgehog pathway.Wiley Interdiscip Rev Dev Biol. 2020 Nov;9(6):e377. doi: 10.1002/wdev.377. Epub 2020 Feb 21. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32084300 Free PMC article. Review.
Cited by
-
Ankmy2 Prevents Smoothened-Independent Hyperactivation of the Hedgehog Pathway via Cilia-Regulated Adenylyl Cyclase Signaling.Dev Cell. 2020 Sep 28;54(6):710-726.e8. doi: 10.1016/j.devcel.2020.06.034. Epub 2020 Jul 22. Dev Cell. 2020. PMID: 32702291 Free PMC article.
-
Rab34 GTPase mediates ciliary membrane formation in the intracellular ciliogenesis pathway.Curr Biol. 2021 Jul 12;31(13):2895-2905.e7. doi: 10.1016/j.cub.2021.04.075. Epub 2021 May 13. Curr Biol. 2021. PMID: 33989527 Free PMC article.
-
The Proteasome and Cul3-Dependent Protein Ubiquitination Is Required for Gli Protein-Mediated Activation of Gene Expression in the Hedgehog Pathway.Cells. 2024 Sep 6;13(17):1496. doi: 10.3390/cells13171496. Cells. 2024. PMID: 39273066 Free PMC article.
-
A comprehensive analysis of Rab GTPases reveals a role for Rab34 in serum starvation-induced primary ciliogenesis.J Biol Chem. 2020 Sep 4;295(36):12674-12685. doi: 10.1074/jbc.RA119.012233. Epub 2020 Jul 15. J Biol Chem. 2020. PMID: 32669361 Free PMC article.
-
Rab34 small GTPase is required for Hedgehog signaling and an early step of ciliary vesicle formation in mouse.J Cell Sci. 2018 Nov 8;131(21):jcs213710. doi: 10.1242/jcs.213710. J Cell Sci. 2018. PMID: 30301781 Free PMC article.
References
-
- Briscoe J., Thérond P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. - PubMed
-
- Cohen M., Briscoe J., Blassberg R. Morphogen interpretation: the transcriptional logic of neural tube patterning. Curr. Opin. Genet. Dev. 2013;23:423–428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

