The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer
- PMID: 29268796
- PMCID: PMC5740748
- DOI: 10.1186/s13073-017-0500-7
The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer
Abstract
Background: Despite a myriad of attempts in the last three decades to diagnose ovarian cancer (OC) earlier, this clinical aim still remains a significant challenge. Aberrant methylation patterns of linked CpGs analyzed in DNA fragments shed by cancers into the bloodstream (i.e. cell-free DNA) can provide highly specific signals indicating cancer presence.
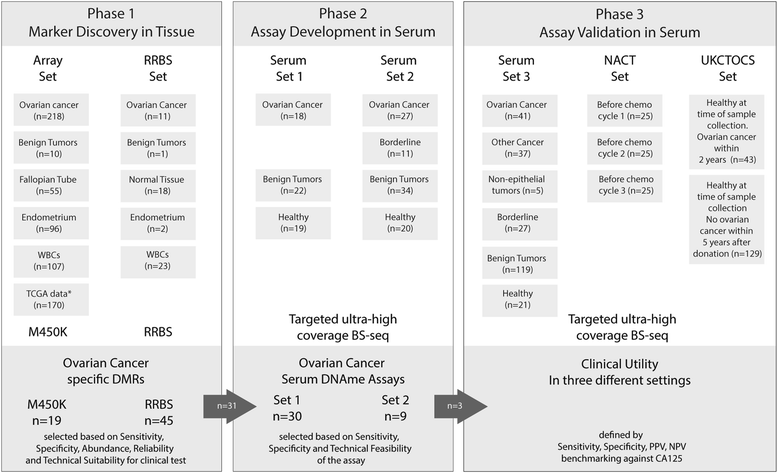
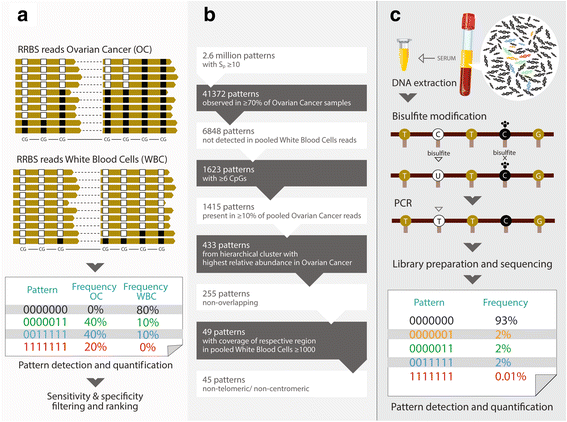
Methods: We analyzed 699 cancerous and non-cancerous tissues using a methylation array or reduced representation bisulfite sequencing to discover the most specific OC methylation patterns. A three-DNA-methylation-serum-marker panel was developed using targeted ultra-high coverage bisulfite sequencing in 151 women and validated in 250 women with various conditions, particularly in those associated with high CA125 levels (endometriosis and other benign pelvic masses), serial samples from 25 patients undergoing neoadjuvant chemotherapy, and a nested case control study of 172 UKCTOCS control arm participants which included serum samples up to two years before OC diagnosis.
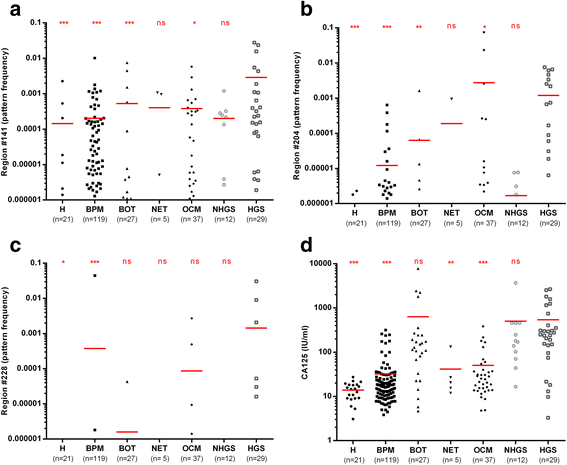
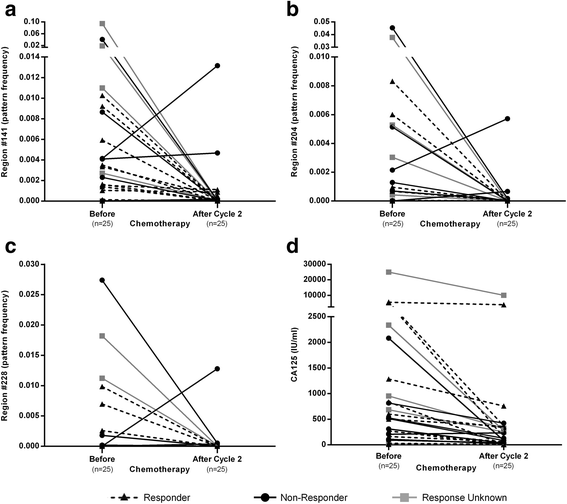
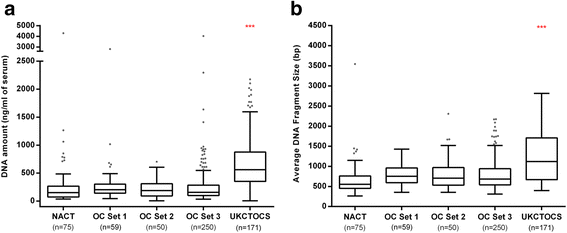
Results: The cell-free DNA amount and average fragment size in the serum samples was up to ten times higher than average published values (based on samples that were immediately processed) due to leakage of DNA from white blood cells owing to delayed time to serum separation. Despite this, the marker panel discriminated high grade serous OC patients from healthy women or patients with a benign pelvic mass with specificity/sensitivity of 90.7% (95% confidence interval [CI] = 84.3-94.8%) and 41.4% (95% CI = 24.1-60.9%), respectively. Levels of all three markers plummeted after exposure to chemotherapy and correctly identified 78% and 86% responders and non-responders (Fisher's exact test, p = 0.04), respectively, which was superior to a CA125 cut-off of 35 IU/mL (20% and 75%). 57.9% (95% CI 34.0-78.9%) of women who developed OC within two years of sample collection were identified with a specificity of 88.1% (95% CI = 77.3-94.3%). Sensitivity and specificity improved further when specifically analyzing CA125 negative samples only (63.6% and 87.5%, respectively).
Conclusions: Our data suggest that DNA methylation patterns in cell-free DNA have the potential to detect a proportion of OCs up to two years in advance of diagnosis and may potentially guide personalized treatment. The prospective use of novel collection vials, which stabilize blood cells and reduce background DNA contamination in serum/plasma samples, will facilitate clinical implementation of liquid biopsy analyses.
Keywords: Cell-free DNA; DNA methylation; Early diagnosis; Ovarian cancer; Personalized treatment; Screening; Serum DNA.
Conflict of interest statement
Ethics approval and consent to participate
Samples were collected prospectively at the University College London Hospital in London and at the Charles University Hospital in Prague and the Department of Gynaecology and Obstetrics. The study was approved by the local research ethics committees: UCL/UCLH Biobank for Studying Health & Disease NC09.13) and the ethics committee of the General University Hospital, Prague approval No.: 22/13 GRANT – 7. RP – EPI-FEM-CARE. All patients provided written informed consent. The UKCTOCS study was approved by the local research ethics committees (UCL/UCLH Biobank for Studying Health & Disease NC09.13) and was approved as part of trial approval by the UK North West Multicentre Research Ethics Committees (North West MREC 00/8/34). All patients provided written informed consent and all studies were conducted in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
HL is an employee and shareholder of Genedata AG. BW is affiliated with a private company. UM has stock ownership in and research funding from Abcodia Pvt Ltd which has an interest in cancer biomarkers. The remaining authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer.Int J Cancer. 2020 Sep 15;147(6):1740-1752. doi: 10.1002/ijc.32984. Epub 2020 Mar 31. Int J Cancer. 2020. PMID: 32191343
-
Methylation patterns in serum DNA for early identification of disseminated breast cancer.Genome Med. 2017 Dec 22;9(1):115. doi: 10.1186/s13073-017-0499-9. Genome Med. 2017. PMID: 29268762 Free PMC article. Clinical Trial.
-
Methylated DNA markers for plasma detection of ovarian cancer: Discovery, validation, and clinical feasibility.Gynecol Oncol. 2022 Jun;165(3):568-576. doi: 10.1016/j.ygyno.2022.03.018. Epub 2022 Mar 31. Gynecol Oncol. 2022. PMID: 35370009 Free PMC article.
-
Early diagnosis of ovarian cancer based on methylation profiles in peripheral blood cell-free DNA: a systematic review.Clin Epigenetics. 2023 Feb 14;15(1):24. doi: 10.1186/s13148-023-01440-w. Clin Epigenetics. 2023. PMID: 36788585 Free PMC article. Review.
-
Diagnostic significance assessment of the circulating cell-free DNA in ovarian cancer: An updated meta-analysis.Gene. 2019 Sep 25;714:143993. doi: 10.1016/j.gene.2019.143993. Epub 2019 Jul 19. Gene. 2019. PMID: 31330238 Review.
Cited by
-
High-throughput approaches for precision medicine in high-grade serous ovarian cancer.J Hematol Oncol. 2020 Oct 9;13(1):134. doi: 10.1186/s13045-020-00971-6. J Hematol Oncol. 2020. PMID: 33036656 Free PMC article. Review.
-
The BRCA1 Methylation and PD-L1 Expression in Sporadic Ovarian Cancer.Int J Gynecol Cancer. 2018 Oct;28(8):1514-1519. doi: 10.1097/IGC.0000000000001334. Int J Gynecol Cancer. 2018. PMID: 30045136 Free PMC article.
-
Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis.EBioMedicine. 2022 May;79:104001. doi: 10.1016/j.ebiom.2022.104001. Epub 2022 Apr 16. EBioMedicine. 2022. PMID: 35439677 Free PMC article. Review.
-
Prognostic Significance of SLFN11 Methylation in Plasma Cell-Free DNA in Advanced High-Grade Serous Ovarian Cancer.Cancers (Basel). 2021 Dec 21;14(1):4. doi: 10.3390/cancers14010004. Cancers (Basel). 2021. PMID: 35008168 Free PMC article.
-
Reducing Ovarian Cancer Mortality Through Early Detection: Approaches Using Circulating Biomarkers.Cancer Prev Res (Phila). 2020 Mar;13(3):241-252. doi: 10.1158/1940-6207.CAPR-19-0184. Cancer Prev Res (Phila). 2020. PMID: 32132118 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- G9901012/MRC_/Medical Research Council/United Kingdom
- C1479/A2884/CRUK_/Cancer Research UK/United Kingdom
- 305428/FP7 Ideas: European Research Council/International
- UCL/UCLH Biomedical Research Centre/National Institute for Health Research/International
- G0801228/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

