Human Cytomegalovirus Tegument Protein pp65 (pUL83) Dampens Type I Interferon Production by Inactivating the DNA Sensor cGAS without Affecting STING
- PMID: 29263269
- PMCID: PMC5827387
- DOI: 10.1128/JVI.01774-17
Human Cytomegalovirus Tegument Protein pp65 (pUL83) Dampens Type I Interferon Production by Inactivating the DNA Sensor cGAS without Affecting STING
Abstract
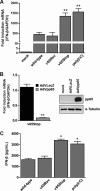
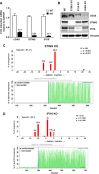
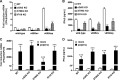
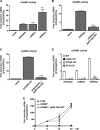
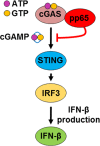
The innate immune response plays a pivotal role during human cytomegalovirus (HCMV) primary infection. Indeed, HCMV infection of primary fibroblasts rapidly triggers strong induction of type I interferons (IFN-I), accompanied by proinflammatory cytokine release. Here, we show that primary human foreskin fibroblasts (HFFs) infected with a mutant HCMV TB40/E strain unable to express UL83-encoded pp65 (v65Stop) produce significantly higher IFN-β levels than HFFs infected with the wild-type TB40/E strain or the pp65 revertant (v65Rev), suggesting that the tegument protein pp65 may dampen IFN-β production. To clarify the mechanisms through which pp65 inhibits IFN-β production, we analyzed the activation of the cGAS/STING/IRF3 axis in HFFs infected with either the wild type, the revertant v65Rev, or the pp65-deficient mutant v65Stop. We found that pp65 selectively binds to cGAS and prevents its interaction with STING, thus inactivating the signaling pathway through the cGAS/STING/IRF3 axis. Consistently, addition of exogenous cGAMP to v65Rev-infected cells triggered the production of IFN-β levels similar to those observed with v65Stop-infected cells, confirming that pp65 inactivation of IFN-β production occurs at the cGAS level. Notably, within the first 24 h of HCMV infection, STING undergoes proteasome degradation independently of the presence or absence of pp65. Collectively, our data provide mechanistic insights into the interplay between HCMV pp65 and cGAS, leading to subsequent immune evasion by this prominent DNA virus.IMPORTANCE Primary human foreskin fibroblasts (HFFs) produce type I IFN (IFN-I) when infected with HCMV. However, we observed significantly higher IFN-β levels when HFFs were infected with HCMV that was unable to express UL83-encoded pp65 (v65Stop), suggesting that pp65 (pUL83) may constitute a viral evasion factor. This study demonstrates that the HCMV tegument protein pp65 inhibits IFN-β production by binding and inactivating cGAS early during infection. In addition, this inhibitory activity specifically targets cGAS, since it can be bypassed via the addition of exogenous cGAMP, even in the presence of pp65. Notably, STING proteasome-mediated degradation was observed in both the presence and absence of pp65. Collectively, our data underscore the important role of the tegument protein pp65 as a critical molecular hub in HCMV's evasion strategy against the innate immune response.
Keywords: IFI16; STING; cGAS; human cytomegalovirus; innate immunity; interactome; interferons; pp65.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
The Viral Tegument Protein pp65 Impairs Transcriptional Upregulation of IL-1β by Human Cytomegalovirus through Inhibition of NF-kB Activity.Viruses. 2018 Oct 16;10(10):567. doi: 10.3390/v10100567. Viruses. 2018. PMID: 30332797 Free PMC article.
-
Regulatory Interaction between the Cellular Restriction Factor IFI16 and Viral pp65 (pUL83) Modulates Viral Gene Expression and IFI16 Protein Stability.J Virol. 2016 Aug 26;90(18):8238-50. doi: 10.1128/JVI.00923-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27384655 Free PMC article.
-
Human Cytomegalovirus Protein UL31 Inhibits DNA Sensing of cGAS to Mediate Immune Evasion.Cell Host Microbe. 2018 Jul 11;24(1):69-80.e4. doi: 10.1016/j.chom.2018.05.007. Epub 2018 Jun 21. Cell Host Microbe. 2018. PMID: 29937271
-
The human cytomegalovirus tegument protein pp65 (pUL83): a key player in innate immune evasion.New Microbiol. 2018 Apr;41(2):87-94. Epub 2018 Jan 31. New Microbiol. 2018. PMID: 29384558 Review.
-
Multifaceted evasion of the interferon response by cytomegalovirus.J Interferon Cytokine Res. 2009 Sep;29(9):609-19. doi: 10.1089/jir.2009.0064. J Interferon Cytokine Res. 2009. PMID: 19708810 Free PMC article. Review.
Cited by
-
The battle between host antiviral innate immunity and immune evasion by cytomegalovirus.Cell Mol Life Sci. 2024 Aug 9;81(1):341. doi: 10.1007/s00018-024-05369-y. Cell Mol Life Sci. 2024. PMID: 39120730 Free PMC article. Review.
-
Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation.Cells. 2022 Sep 28;11(19):3043. doi: 10.3390/cells11193043. Cells. 2022. PMID: 36231006 Free PMC article. Review.
-
Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity.J Virol. 2018 Jul 17;92(15):e00841-18. doi: 10.1128/JVI.00841-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29793952 Free PMC article.
-
Camouflage and interception: how pathogens evade detection by intracellular nucleic acid sensors.Immunology. 2019 Mar;156(3):217-227. doi: 10.1111/imm.13030. Epub 2018 Dec 18. Immunology. 2019. PMID: 30499584 Free PMC article. Review.
-
Subviral Dense Bodies of Human Cytomegalovirus Induce an Antiviral Type I Interferon Response.Cells. 2022 Dec 13;11(24):4028. doi: 10.3390/cells11244028. Cells. 2022. PMID: 36552792 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

