pKID Binds to KIX via an Unstructured Transition State with Nonnative Interactions
- PMID: 29262364
- PMCID: PMC5770965
- DOI: 10.1016/j.bpj.2017.10.016
pKID Binds to KIX via an Unstructured Transition State with Nonnative Interactions
Abstract
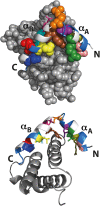
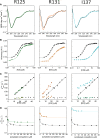
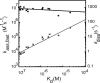
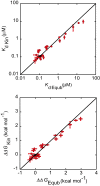
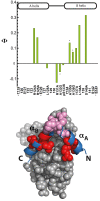
Understanding the detailed mechanism of interaction of intrinsically disordered proteins with their partners is crucial to comprehend their functions in signaling and transcription. Through its interaction with KIX, the disordered pKID region of CREB protein is central in the transcription of cAMP responsive genes, including those involved in long-term memory. Numerous simulation studies have investigated these interactions. Combined with experimental results, these can provide valuable and comprehensive understanding of the mechanisms involved. Here, we probe the transition state of this interaction experimentally through analyzing the kinetic effect of mutating both interface and solvent exposed residues in pKID. We show that very few specific interactions between pKID and KIX are required in the initial binding process. Only a small number of weak interactions are formed at the transition state, including nonnative interactions, and most of the folding occurs after the initial binding event. These properties are consistent with computational results and also the majority of experimental studies of intrinsically disordered protein coupled folding and binding in other protein systems, suggesting that these may be common features.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):9614-9. doi: 10.1073/pnas.1512799112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195786 Free PMC article.
-
Phosphorylation of the IDP KID Modulates Affinity for KIX by Increasing the Lifetime of the Complex.Biophys J. 2017 Dec 19;113(12):2706-2712. doi: 10.1016/j.bpj.2017.10.015. Biophys J. 2017. PMID: 29262363 Free PMC article.
-
Mechanism of pKID/KIX Association Studied by Molecular Dynamics Free Energy Simulations.J Phys Chem B. 2016 Aug 25;120(33):8186-92. doi: 10.1021/acs.jpcb.6b01792. Epub 2016 Apr 20. J Phys Chem B. 2016. PMID: 27054660
-
Insights into Coupled Folding and Binding Mechanisms from Kinetic Studies.J Biol Chem. 2016 Mar 25;291(13):6689-95. doi: 10.1074/jbc.R115.692715. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851275 Free PMC article. Review.
-
Dynamic conformational flexibility and molecular interactions of intrinsically disordered proteins.J Biosci. 2020;45:29. J Biosci. 2020. PMID: 32020911 Review.
Cited by
-
A disordered encounter complex is central to the yeast Abp1p SH3 domain binding pathway.PLoS Comput Biol. 2020 Sep 14;16(9):e1007815. doi: 10.1371/journal.pcbi.1007815. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32925900 Free PMC article.
-
Residual Structure Accelerates Binding of Intrinsically Disordered ACTR by Promoting Efficient Folding upon Encounter.J Mol Biol. 2019 Jan 18;431(2):422-432. doi: 10.1016/j.jmb.2018.12.001. Epub 2018 Dec 7. J Mol Biol. 2019. PMID: 30528464 Free PMC article.
-
Position-, disorder-, and salt-dependent diffusion in binding-coupled-folding of intrinsically disordered proteins.Phys Chem Chem Phys. 2019 Mar 6;21(10):5634-5645. doi: 10.1039/c8cp06803h. Phys Chem Chem Phys. 2019. PMID: 30793144 Free PMC article.
-
Designed Mutations Alter the Binding Pathways of an Intrinsically Disordered Protein.Sci Rep. 2019 Apr 16;9(1):6172. doi: 10.1038/s41598-019-42717-6. Sci Rep. 2019. PMID: 30992509 Free PMC article.
-
Understanding the Binding Induced Folding of Intrinsically Disordered Proteins by Protein Engineering: Caveats and Pitfalls.Int J Mol Sci. 2020 May 15;21(10):3484. doi: 10.3390/ijms21103484. Int J Mol Sci. 2020. PMID: 32429036 Free PMC article. Review.
References
-
- Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

