TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase
- PMID: 29253195
- PMCID: PMC5778498
- DOI: 10.1093/nar/gkx1247
TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase
Abstract
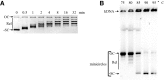
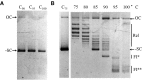
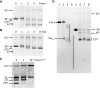
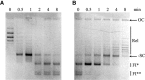
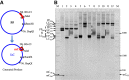
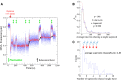
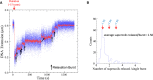
DNA topoisomerases are essential enzymes involved in all the DNA processes and among them, type IA topoisomerases emerged as a key actor in the maintenance of genome stability. The hyperthermophilic archaeon, Sulfolobus solfataricus, contains three topoisomerases IA including one classical named TopA. SsoTopA is very efficient at unlinking DNA catenanes, grouping SsoTopA into the topoisomerase III family. SsoTopA is active over a wide range of temperatures and at temperatures of up to 85°C it produces highly unwound DNA. At higher temperatures, SsoTopA unlinks the two DNA strands. Thus depending on the temperature, SsoTopA is able to either prevent or favor DNA melting. While canonical topoisomerases III require a single-stranded DNA region or a nick in one of the circles to decatenate them, we show for the first time that a type I topoisomerase, SsoTopA, is able to efficiently unlink covalently closed catenanes, with no additional partners. By using single molecule experiments we demonstrate that SsoTopA requires the presence of a short single-stranded DNA region to be efficient. The unexpected decatenation property of SsoTopA probably comes from its high ability to capture this unwound region. This points out a possible role of TopA in S. solfataricus as a decatenase in Sulfolobus.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Oligonucleotide cleavage and rejoining by topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus: temperature dependence and strand annealing-promoted DNA religation.Mol Microbiol. 2006 May;60(3):783-94. doi: 10.1111/j.1365-2958.2006.05133.x. Mol Microbiol. 2006. PMID: 16629677
-
DNA topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus with specific DNA cleavage activity.J Bacteriol. 2003 Sep;185(18):5500-7. doi: 10.1128/JB.185.18.5500-5507.2003. J Bacteriol. 2003. PMID: 12949102 Free PMC article.
-
Single-molecule analysis uncovers the difference between the kinetics of DNA decatenation by bacterial topoisomerases I and III.Nucleic Acids Res. 2014 Oct;42(18):11657-67. doi: 10.1093/nar/gku785. Epub 2014 Sep 17. Nucleic Acids Res. 2014. PMID: 25232096 Free PMC article.
-
The chromosome replication machinery of the archaeon Sulfolobus solfataricus.J Biol Chem. 2006 Jun 2;281(22):15029-32. doi: 10.1074/jbc.R500029200. Epub 2006 Feb 8. J Biol Chem. 2006. PMID: 16467299 Review.
-
Structure and mechanism of action of type IA DNA topoisomerases.Biochemistry (Mosc). 2009 Dec;74(13):1467-81. doi: 10.1134/s0006297909130045. Biochemistry (Mosc). 2009. PMID: 20210704 Review.
Cited by
-
What's on the Other Side of the Gate: A Structural Perspective on DNA Gate Opening of Type IA and IIA DNA Topoisomerases.Int J Mol Sci. 2023 Feb 16;24(4):3986. doi: 10.3390/ijms24043986. Int J Mol Sci. 2023. PMID: 36835394 Free PMC article. Review.
-
BIOPHYSICS MEETS GENE THERAPY: HOW EXPLORING SUPERCOILING-DEPENDENT STRUCTURAL CHANGES IN DNA LED TO THE DEVELOPMENT OF MINIVECTOR DNA.Technol Innov. 2019 Aug;20(4):427-439. doi: 10.21300/20.4.2019.427. Epub 2019 Aug 1. Technol Innov. 2019. PMID: 33815681 Free PMC article.
-
Structural and biochemical basis for DNA and RNA catalysis by human Topoisomerase 3β.Nat Commun. 2022 Aug 9;13(1):4656. doi: 10.1038/s41467-022-32221-3. Nat Commun. 2022. PMID: 35945419 Free PMC article.
-
Topoisomerase VI is a chirally-selective, preferential DNA decatenase.Elife. 2022 Jan 25;11:e67021. doi: 10.7554/eLife.67021. Elife. 2022. PMID: 35076393 Free PMC article.
-
The many lives of type IA topoisomerases.J Biol Chem. 2020 May 15;295(20):7138-7153. doi: 10.1074/jbc.REV120.008286. Epub 2020 Apr 10. J Biol Chem. 2020. PMID: 32277049 Free PMC article. Review.
References
-
- Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001; 70:369–413. - PubMed
-
- Wang J.C. Cellular roles of dna topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. - PubMed
-
- Duguet M., Serre M.-C., Bouthier de La Tour C.. A universal type IA topoisomerase fold. J. Mol. Biol. 2006; 359:805–812. - PubMed
-
- Viard T., Bouthier de La Tour C.. Type IA topoisomerases: a simple puzzle. Biochimie. 2007; 89:456–467. - PubMed
-
- Tse-Dinh Y.C. Bacterial and archeal type I topoisomerases. Biochim. Biophys. Acta. 1998; 1400:19–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

