Zoonotic origin and transmission of Middle East respiratory syndrome coronavirus in the UAE
- PMID: 29239118
- PMCID: PMC5893383
- DOI: 10.1111/zph.12435
Zoonotic origin and transmission of Middle East respiratory syndrome coronavirus in the UAE
Abstract
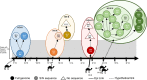
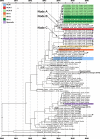
Since the emergence of Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, there have been a number of clusters of human-to-human transmission. These cases of human-to-human transmission involve close contact and have occurred primarily in healthcare settings, and they are suspected to result from repeated zoonotic introductions. In this study, we sequenced whole MERS-CoV genomes directly from respiratory samples collected from 23 confirmed MERS cases in the United Arab Emirates (UAE). These samples included cases from three nosocomial and three household clusters. The sequences were analysed for changes and relatedness with regard to the collected epidemiological data and other available MERS-CoV genomic data. Sequence analysis supports the epidemiological data within the clusters, and further, suggests that these clusters emerged independently. To understand how and when these clusters emerged, respiratory samples were taken from dromedary camels, a known host of MERS-CoV, in the same geographic regions as the human clusters. Middle East respiratory syndrome coronavirus genomes from six virus-positive animals were sequenced, and these genomes were nearly identical to those found in human patients from corresponding regions. These data demonstrate a genetic link for each of these clusters to a camel and support the hypothesis that human MERS-CoV diversity results from multiple zoonotic introductions.
Keywords: dromedary camel; epidemiology; genomics; middle east respiratory syndrome; viral pathogens; zoonoses.
© 2017 The Authors. Zoonoses and Public Health Published by Blackwell Verlag GmbH.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



Similar articles
-
Limited Genetic Diversity Detected in Middle East Respiratory Syndrome-Related Coronavirus Variants Circulating in Dromedary Camels in Jordan.Viruses. 2021 Mar 31;13(4):592. doi: 10.3390/v13040592. Viruses. 2021. PMID: 33807288 Free PMC article.
-
Epidemiological investigation of Middle East respiratory syndrome coronavirus in dromedary camel farms linked with human infection in Abu Dhabi Emirate, United Arab Emirates.Virus Genes. 2016 Dec;52(6):848-854. doi: 10.1007/s11262-016-1367-1. Epub 2016 Jun 29. Virus Genes. 2016. PMID: 27357298 Free PMC article.
-
Diversity of Middle East respiratory syndrome coronaviruses in 109 dromedary camels based on full-genome sequencing, Abu Dhabi, United Arab Emirates.Emerg Microbes Infect. 2017 Nov 8;6(11):e101. doi: 10.1038/emi.2017.89. Emerg Microbes Infect. 2017. PMID: 29116217 Free PMC article.
-
Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction.Pathog Glob Health. 2015;109(8):354-62. doi: 10.1080/20477724.2015.1122852. Pathog Glob Health. 2015. PMID: 26924345 Free PMC article. Review.
-
Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir.Virol J. 2016 Jun 3;13:87. doi: 10.1186/s12985-016-0544-0. Virol J. 2016. PMID: 27255185 Free PMC article. Review.
Cited by
-
Middle East Respiratory Syndrome Coronavirus Could be a Priority Pathogen to Cause Public Health Emergency: Noticeable Features and Counteractive Measures.Environ Health Insights. 2024 Aug 15;18:11786302241271545. doi: 10.1177/11786302241271545. eCollection 2024. Environ Health Insights. 2024. PMID: 39156879 Free PMC article.
-
Interrogating the lack of diversity of thought in the pandemic response that led to mistakes - holistic evidence-based approach to deal with future pandemics.Front Public Health. 2023 Dec 21;11:1310210. doi: 10.3389/fpubh.2023.1310210. eCollection 2023. Front Public Health. 2023. PMID: 38192553 Free PMC article.
-
Zoonotic diseases transmitted from the camels.Front Vet Sci. 2023 Oct 19;10:1244833. doi: 10.3389/fvets.2023.1244833. eCollection 2023. Front Vet Sci. 2023. PMID: 37929289 Free PMC article. Review.
-
Isolation and genetic characterization of MERS-CoV from dromedary camels in the United Arab Emirates.Front Vet Sci. 2023 Aug 31;10:1182165. doi: 10.3389/fvets.2023.1182165. eCollection 2023. Front Vet Sci. 2023. PMID: 37720473 Free PMC article.
-
The Protective Efficacy of Single-Dose Nasal Immunization with Cold-Adapted Live-Attenuated MERS-CoV Vaccine against Lethal MERS-CoV Infections in Mice.Vaccines (Basel). 2023 Aug 10;11(8):1353. doi: 10.3390/vaccines11081353. Vaccines (Basel). 2023. PMID: 37631921 Free PMC article.
References
-
- Adney, D. R. , van Doremalen, N. , Brown, V. R. , Bushmaker, T. , Scott, D. , de Wit, E. , … Munster, V. J. (2014). Replication and shedding of MERS‐CoV in upper respiratory tract of inoculated dromedary camels. Emerging Infectious Diseases, 20(12), 1999–2005. 10.3201/eid2012.141280 - DOI - PMC - PubMed
-
- Al Hammadi, Z. M. , Chu, D. K. , Eltahir, Y. M. , Al Hosani, F. , Al Mulla, M. , Tarnini, W. , … Poon, L. L. (2015). Asymptomatic MERS‐CoV infection in humans possibly linked to infected dromedaries imported from Oman to United Arab Emirates, May 2015. Emerging Infectious Diseases, 21(12), 2197–2200. 10.3201/eid2112.151132 - DOI - PMC - PubMed
-
- Al Hosani, F. I. , Pringle, K. , Al Mulla, M. , Kim, L. , Pham, H. , Alami, N. N. , … Gerber, S. I. (2016). Response to emergence of Middle East respiratory syndrome coronavirus, Abu Dhabi, United Arab Emirates, 2013‐2014. Emerging Infectious Diseases, 22(7), 1162–1168. 10.3201/eid2207.160040 - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

