Human Cytomegalovirus UL111A and US27 Gene Products Enhance the CXCL12/CXCR4 Signaling Axis via Distinct Mechanisms
- PMID: 29237840
- PMCID: PMC5809719
- DOI: 10.1128/JVI.01981-17
Human Cytomegalovirus UL111A and US27 Gene Products Enhance the CXCL12/CXCR4 Signaling Axis via Distinct Mechanisms
Abstract
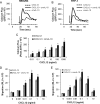
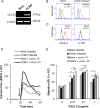
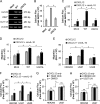
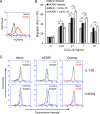
Human cytomegalovirus (HCMV) is a prevalent pathogen that establishes lifelong infection in the host. Virus persistence is aided by extensive manipulation of the host immune system, particularly cytokine and chemokine signaling pathways. The HCMV UL111A gene encodes cmvIL-10, an ortholog of human interleukin-10 that has many immunomodulatory effects. We found that cmvIL-10 increased signaling outcomes from human CXCR4, a chemokine receptor with essential roles in hematopoiesis and immune cell trafficking, in response to its natural ligand CXCL12. Calcium flux and chemotaxis to CXCL12 were significantly greater in the presence of cmvIL-10 in monocytes, epithelial cells, and fibroblasts that express CXCR4. cmvIL-10 effects on CXCL12/CXCR4 signaling required the IL-10 receptor and Stat3 activation. Heightened signaling occurred both in HCMV-infected cells and in uninfected bystander cells, suggesting that cmvIL-10 may broadly influence chemokine networks by paracrine signaling during infection. Moreover, CXCL12/CXCR4 signaling was amplified in HCMV-infected cells compared to mock-infected cells even in the absence of cmvIL-10. Enhanced CXCL12/CXCR4 outcomes were associated with expression of the virally encoded chemokine receptor US27, and CXCL12/CXCR4 activation was reduced in cells infected with a deletion mutant lacking US27 (TB40/E-mCherry-US27Δ). US27 effects were Stat3 independent but required close proximity to CXCR4 in cell membranes of either HCMV-infected or US27-transfected cells. Thus, HCMV encodes two proteins, cmvIL-10 and US27, that exhibit distinct mechanisms for enhancing CXCR4 signaling. Either individually or in combination, cmvIL-10 and US27 may enable HCMV to exquisitely manipulate CXCR4 signaling to alter host immune responses and modify cell trafficking patterns during infection.IMPORTANCE The human chemokine system plays a central role in host defense, as evidenced by the many strategies devised by viruses for manipulating it. Human cytomegalovirus (HCMV) is widespread in the human population, but infection rarely causes disease except in immunocompromised hosts. We found that two different HCMV proteins, cmvIL-10 and US27, act through distinct mechanisms to upregulate the signaling activity of a cellular chemokine receptor, CXCR4. cmvIL-10 is a secreted viral cytokine that affects CXCR4 signaling in both infected and uninfected cells, while US27 is a component of the virus particle and impacts CXCR4 activity only in infected cells. Both cmvIL-10 and US27 promote increased intracellular calcium signaling and cell migration in response to chemokine CXCL12 binding to CXCR4. Our results demonstrate that HCMV exerts fine control over the CXCL12/CXCR4 pathway, which could lead to enhanced virus dissemination, altered immune cell trafficking, and serious health implications for HCMV patients.
Keywords: CMV; CXCL12; CXCR4; US27; chemokine; chemokine receptors; cmvIL-10; cytokine; cytomegalovirus.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
Identification of a novel signaling complex containing host chemokine receptor CXCR4, Interleukin-10 receptor, and human cytomegalovirus US27.Virology. 2020 Sep;548:49-58. doi: 10.1016/j.virol.2020.06.006. Epub 2020 Jun 17. Virology. 2020. PMID: 32838946 Free PMC article.
-
The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through Gβγ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1.J Virol. 2018 Nov 12;92(23):e00644-18. doi: 10.1128/JVI.00644-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209167 Free PMC article.
-
Effect of human cytomegalovirus (HCMV) US27 on CXCR4 receptor internalization measured by fluorogen-activating protein (FAP) biosensors.PLoS One. 2017 Feb 16;12(2):e0172042. doi: 10.1371/journal.pone.0172042. eCollection 2017. PLoS One. 2017. PMID: 28207860 Free PMC article.
-
The human cytomegalovirus chemokine receptor homolog encoded by US27.Virus Genes. 2017 Aug;53(4):516-521. doi: 10.1007/s11262-017-1462-y. Epub 2017 Apr 26. Virus Genes. 2017. PMID: 28447191 Review.
-
Viral chemokine receptors and chemokines in human cytomegalovirus trafficking and interaction with the immune system. CMV chemokine receptors.Curr Top Microbiol Immunol. 2002;269:203-34. doi: 10.1007/978-3-642-59421-2_13. Curr Top Microbiol Immunol. 2002. PMID: 12224510 Review.
Cited by
-
The Role of CMV Infection in Primary Lesions, Development and Clinical Expression of Atherosclerosis.J Clin Med. 2022 Jul 1;11(13):3832. doi: 10.3390/jcm11133832. J Clin Med. 2022. PMID: 35807114 Free PMC article. Review.
-
Human Cytomegalovirus Primary Infection and Reactivation: Insights From Virion-Carried Molecules.Front Microbiol. 2020 Jul 14;11:1511. doi: 10.3389/fmicb.2020.01511. eCollection 2020. Front Microbiol. 2020. PMID: 32765441 Free PMC article. Review.
-
Risk prediction of CMV reactivation after allogeneic stem cell transplantation using five non-HLA immunogenetic polymorphisms.Ann Hematol. 2022 Jul;101(7):1567-1576. doi: 10.1007/s00277-022-04841-8. Epub 2022 May 7. Ann Hematol. 2022. PMID: 35525883 Free PMC article.
-
Identification of a novel signaling complex containing host chemokine receptor CXCR4, Interleukin-10 receptor, and human cytomegalovirus US27.Virology. 2020 Sep;548:49-58. doi: 10.1016/j.virol.2020.06.006. Epub 2020 Jun 17. Virology. 2020. PMID: 32838946 Free PMC article.
-
Challenging the Conventional Interpretation of HCMV Seronegativity.Microorganisms. 2021 Nov 18;9(11):2382. doi: 10.3390/microorganisms9112382. Microorganisms. 2021. PMID: 34835508 Free PMC article.
References
-
- Azevedo LS, Pierrotti LC, Abdala E, Costa SF, Strabelli TM, Campos SV, Ramos JF, Latif AZ, Litvinov N, Maluf NZ, Caiaffa Filho HH, Pannuti CS, Lopes MH, Santos VA, Linardi Cda C, Yasuda MA, Marques HH. 2015. Cytomegalovirus infection in transplant recipients. Clinics (Sao Paulo) 70:515–523. doi:10.6061/clinics/2015(07)09. - DOI - PMC - PubMed
-
- Spencer JV. 2012. Trojan horses and fake immunity idols: molecular mimicry of host immune mediators by human cytomegalovirus, p 41–64. In Tyring S, Magel GD (ed), Herpesviridae: a look into this unique family of viruses. InTech, London, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

