Effector CD8 T cells dedifferentiate into long-lived memory cells
- PMID: 29236683
- PMCID: PMC5965677
- DOI: 10.1038/nature25144
Effector CD8 T cells dedifferentiate into long-lived memory cells
Abstract
Memory CD8 T cells that circulate in the blood and are present in lymphoid organs are an essential component of long-lived T cell immunity. These memory CD8 T cells remain poised to rapidly elaborate effector functions upon re-exposure to pathogens, but also have many properties in common with naive cells, including pluripotency and the ability to migrate to the lymph nodes and spleen. Thus, memory cells embody features of both naive and effector cells, fuelling a long-standing debate centred on whether memory T cells develop from effector cells or directly from naive cells. Here we show that long-lived memory CD8 T cells are derived from a subset of effector T cells through a process of dedifferentiation. To assess the developmental origin of memory CD8 T cells, we investigated changes in DNA methylation programming at naive and effector cell-associated genes in virus-specific CD8 T cells during acute lymphocytic choriomeningitis virus infection in mice. Methylation profiling of terminal effector versus memory-precursor CD8 T cell subsets showed that, rather than retaining a naive epigenetic state, the subset of cells that gives rise to memory cells acquired de novo DNA methylation programs at naive-associated genes and became demethylated at the loci of classically defined effector molecules. Conditional deletion of the de novo methyltransferase Dnmt3a at an early stage of effector differentiation resulted in reduced methylation and faster re-expression of naive-associated genes, thereby accelerating the development of memory cells. Longitudinal phenotypic and epigenetic characterization of the memory-precursor effector subset of virus-specific CD8 T cells transferred into antigen-free mice revealed that differentiation to memory cells was coupled to erasure of de novo methylation programs and re-expression of naive-associated genes. Thus, epigenetic repression of naive-associated genes in effector CD8 T cells can be reversed in cells that develop into long-lived memory CD8 T cells while key effector genes remain demethylated, demonstrating that memory T cells arise from a subset of fate-permissive effector T cells.
Conflict of interest statement
The authors report no competing financial interests.
Figures

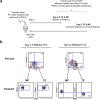



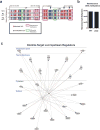


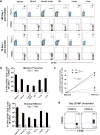
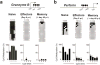
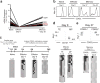

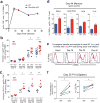
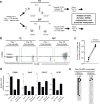
Comment in
-
The origins of memory T cells.Nature. 2017 Dec 21;552(7685):337-339. doi: 10.1038/d41586-017-08280-8. Nature. 2017. PMID: 29293221 No abstract available.
Similar articles
-
DNA methylation by DNA methyltransferase 1 is critical for effector CD8 T cell expansion.J Immunol. 2006 Apr 15;176(8):4562-72. doi: 10.4049/jimmunol.176.8.4562. J Immunol. 2006. PMID: 16585546
-
De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation.Proc Natl Acad Sci U S A. 2016 Sep 20;113(38):10631-6. doi: 10.1073/pnas.1524490113. Epub 2016 Aug 31. Proc Natl Acad Sci U S A. 2016. PMID: 27582468 Free PMC article.
-
Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis.J Exp Med. 2017 Jun 5;214(6):1593-1606. doi: 10.1084/jem.20161760. Epub 2017 May 10. J Exp Med. 2017. PMID: 28490440 Free PMC article.
-
MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections.Blood. 2013 May 30;121(22):4473-83. doi: 10.1182/blood-2012-06-435412. Epub 2013 Apr 17. Blood. 2013. PMID: 23596046
-
Generating long-lived CD8(+) T-cell memory: Insights from epigenetic programs.Eur J Immunol. 2016 Jul;46(7):1548-62. doi: 10.1002/eji.201545550. Eur J Immunol. 2016. PMID: 27230488 Free PMC article. Review.
Cited by
-
Decitabine-Mediated Epigenetic Reprograming Enhances Anti-leukemia Efficacy of CD123-Targeted Chimeric Antigen Receptor T-Cells.Front Immunol. 2020 Aug 18;11:1787. doi: 10.3389/fimmu.2020.01787. eCollection 2020. Front Immunol. 2020. PMID: 32973749 Free PMC article.
-
Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25667-25678. doi: 10.1073/pnas.2008571117. Epub 2020 Sep 25. Proc Natl Acad Sci U S A. 2020. PMID: 32978300 Free PMC article.
-
Rewriting History: Epigenetic Reprogramming of CD8+ T Cell Differentiation to Enhance Immunotherapy.Trends Immunol. 2020 Aug;41(8):665-675. doi: 10.1016/j.it.2020.06.008. Epub 2020 Jul 2. Trends Immunol. 2020. PMID: 32624330 Free PMC article. Review.
-
IL-2/IL-7-inducible factors pioneer the path to T cell differentiation in advance of lineage-defining factors.EMBO J. 2020 Nov 16;39(22):e105220. doi: 10.15252/embj.2020105220. Epub 2020 Sep 15. EMBO J. 2020. PMID: 32930455 Free PMC article.
-
Chromatin accessibility profiling methods.Nat Rev Methods Primers. 2021;1:10. doi: 10.1038/s43586-020-00008-9. Epub 2021 Jan 21. Nat Rev Methods Primers. 2021. PMID: 38410680 Free PMC article.
References
-
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

