Pandemic H1N1 influenza A viruses suppress immunogenic RIPK3-driven dendritic cell death
- PMID: 29203926
- PMCID: PMC5715119
- DOI: 10.1038/s41467-017-02035-9
Pandemic H1N1 influenza A viruses suppress immunogenic RIPK3-driven dendritic cell death
Abstract
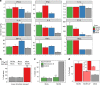
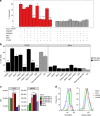
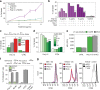
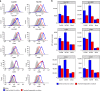
The risk of emerging pandemic influenza A viruses (IAVs) that approach the devastating 1918 strain motivates finding strain-specific host-pathogen mechanisms. During infection, dendritic cells (DC) mature into antigen-presenting cells that activate T cells, linking innate to adaptive immunity. DC infection with seasonal IAVs, but not with the 1918 and 2009 pandemic strains, induces global RNA degradation. Here, we show that DC infection with seasonal IAV causes immunogenic RIPK3-mediated cell death. Pandemic IAV suppresses this immunogenic DC cell death. Only DC infected with seasonal IAV, but not with pandemic IAV, enhance maturation of uninfected DC and T cell proliferation. In vivo, circulating T cell levels are reduced after pandemic, but not seasonal, IAV infection. Using recombinant viruses, we identify the HA genomic segment as the mediator of cell death inhibition. These results show how pandemic influenza viruses subvert the immune response.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Nucleotide oligomerization and binding domain 2-dependent dendritic cell activation is necessary for innate immunity and optimal CD8+ T Cell responses to influenza A virus infection.J Virol. 2014 Aug;88(16):8946-55. doi: 10.1128/JVI.01110-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872587 Free PMC article.
-
Human Dendritic Cell Response Signatures Distinguish 1918, Pandemic, and Seasonal H1N1 Influenza Viruses.J Virol. 2015 Oct;89(20):10190-205. doi: 10.1128/JVI.01523-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223639 Free PMC article.
-
Conserved epitopes dominate cross-CD8+ T-cell responses against influenza A H1N1 virus among Asian populations.Eur J Immunol. 2013 Aug;43(8):2055-69. doi: 10.1002/eji.201343417. Epub 2013 Jun 14. Eur J Immunol. 2013. PMID: 23681926 Clinical Trial.
-
Immunity to seasonal and pandemic influenza A viruses.Microbes Infect. 2011 May;13(5):489-501. doi: 10.1016/j.micinf.2011.01.007. Epub 2011 Feb 2. Microbes Infect. 2011. PMID: 21295153 Free PMC article. Review.
-
Dendritic cells and influenza A virus infection.Virulence. 2012 Nov 15;3(7):603-8. doi: 10.4161/viru.21864. Epub 2012 Oct 17. Virulence. 2012. PMID: 23076333 Free PMC article. Review.
Cited by
-
Influenza viral matrix 1 protein aggravates viral pathogenicity by inducing TLR4-mediated reactive oxygen species production and apoptotic cell death.Cell Death Dis. 2023 Mar 30;14(3):228. doi: 10.1038/s41419-023-05749-5. Cell Death Dis. 2023. PMID: 36990977 Free PMC article.
-
M1 Macrophages Are More Susceptible to Necroptosis.J Cell Immunol. 2021;3(2):97-102. doi: 10.33696/immunology.3.084. J Cell Immunol. 2021. PMID: 33959729 Free PMC article.
-
Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1.Cell Death Dis. 2018 Jul 26;9(8):816. doi: 10.1038/s41419-018-0868-3. Cell Death Dis. 2018. PMID: 30050136 Free PMC article.
-
MLKL-Driven Inflammasome Activation and Caspase-8 Mediate Inflammatory Cell Death in Influenza A Virus Infection.mBio. 2023 Apr 25;14(2):e0011023. doi: 10.1128/mbio.00110-23. Epub 2023 Feb 28. mBio. 2023. PMID: 36852999 Free PMC article.
-
Inflammatory cell death: how macrophages sense neighbouring cell infection and damage.Biochem Soc Trans. 2023 Feb 27;51(1):303-313. doi: 10.1042/BST20220807. Biochem Soc Trans. 2023. PMID: 36695550 Free PMC article. Review.
References
-
- Ayllon J, Garcia-Sastre A. The NS1 protein: a multitasking virulence factor. Curr. Top. Microbiol. Immunol. 2015;386:73–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

