Linear Integration of ERK Activity Predominates over Persistence Detection in Fra-1 Regulation
- PMID: 29199017
- PMCID: PMC5746471
- DOI: 10.1016/j.cels.2017.10.019
Linear Integration of ERK Activity Predominates over Persistence Detection in Fra-1 Regulation
Abstract
ERK signaling regulates the expression of target genes, but it is unclear how ERK activity dynamics are interpreted. Here, we investigate this question using simultaneous, live, single-cell imaging of two ERK activity reporters and expression of Fra-1, a target gene controlling epithelial cell identity. We find that Fra-1 is expressed in proportion to the amplitude and duration of ERK activity. In contrast to previous "persistence detector" and "selective filter" models in which Fra-1 expression only occurs when ERK activity persists beyond a threshold duration, our observations demonstrate that the network regulating Fra-1 expression integrates total ERK activity and responds to it linearly. However, exploration of a generalized mathematical model of the Fra-1 coherent feedforward loop demonstrates that it can perform either linear integration or persistence detection, depending on the basal mRNA production rate and protein production delays. Our data indicate that significant basal expression and short delays cause Fra-1 to respond linearly to integrated ERK activity.
Keywords: EGF; EGFR; FOSL1; FRET; KTR; MAPK; Ras; c-Fos degradation; signal transduction dynamics; transcription.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

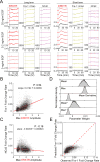
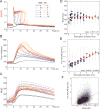

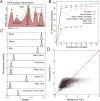
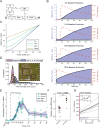
Comment in
-
Decoding Signal Processing at the Single-Cell Level.Cell Syst. 2017 Dec 27;5(6):542-543. doi: 10.1016/j.cels.2017.12.003. Cell Syst. 2017. PMID: 29284127
Similar articles
-
Elevated ERK-MAP kinase activity protects the FOS family member FRA-1 against proteasomal degradation in colon carcinoma cells.J Cell Sci. 2003 Dec 15;116(Pt 24):4957-63. doi: 10.1242/jcs.00812. J Cell Sci. 2003. PMID: 14625389
-
A 19S proteasomal subunit cooperates with an ERK MAPK-regulated degron to regulate accumulation of Fra-1 in tumour cells.Oncogene. 2012 Apr 5;31(14):1817-24. doi: 10.1038/onc.2011.375. Epub 2011 Aug 29. Oncogene. 2012. PMID: 21874050
-
Matrix metalloproteinase/epidermal growth factor receptor/mitogen-activated protein kinase signaling regulate fra-1 induction by cigarette smoke in lung epithelial cells.Am J Respir Cell Mol Biol. 2005 Jan;32(1):72-81. doi: 10.1165/rcmb.2004-0198OC. Epub 2004 Nov 4. Am J Respir Cell Mol Biol. 2005. PMID: 15528491
-
A JNK-independent signaling pathway regulates TNF alpha-stimulated, c-Jun-driven FRA-1 protooncogene transcription in pulmonary epithelial cells.J Immunol. 2006 Nov 15;177(10):7193-202. doi: 10.4049/jimmunol.177.10.7193. J Immunol. 2006. PMID: 17082637
-
c-Jun Is Required for Nuclear Factor-κB-Dependent, LPS-Stimulated Fos-Related Antigen-1 Transcription in Alveolar Macrophages.Am J Respir Cell Mol Biol. 2016 Nov;55(5):667-674. doi: 10.1165/rcmb.2016-0028OC. Am J Respir Cell Mol Biol. 2016. PMID: 27286066 Free PMC article.
Cited by
-
Coupling-dependent metabolic ultradian rhythms in confluent cells.Proc Natl Acad Sci U S A. 2022 Nov 8;119(45):e2211142119. doi: 10.1073/pnas.2211142119. Epub 2022 Nov 2. Proc Natl Acad Sci U S A. 2022. PMID: 36322771 Free PMC article.
-
Re-expression of epigenetically silenced PTPRR by histone acetylation sensitizes RAS-mutant lung adenocarcinoma to SHP2 inhibition.Cell Mol Life Sci. 2024 Jan 28;81(1):64. doi: 10.1007/s00018-023-05034-w. Cell Mol Life Sci. 2024. PMID: 38280930 Free PMC article.
-
A guide to ERK dynamics, part 2: downstream decoding.Biochem J. 2023 Dec 13;480(23):1909-1928. doi: 10.1042/BCJ20230277. Biochem J. 2023. PMID: 38038975 Free PMC article.
-
CODEX, a neural network approach to explore signaling dynamics landscapes.Mol Syst Biol. 2021 Apr;17(4):e10026. doi: 10.15252/msb.202010026. Mol Syst Biol. 2021. PMID: 33835701 Free PMC article.
-
Expression and function of FRA1 protein in tumors.Mol Biol Rep. 2020 Jan;47(1):737-752. doi: 10.1007/s11033-019-05123-9. Epub 2019 Oct 14. Mol Biol Rep. 2020. PMID: 31612408 Review.
References
-
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. - PubMed
-
- Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y, Shionyu C, Matsuda M. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol Cell. 2013;52:529–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

