Multiplex single-cell visualization of nucleic acids and protein during HIV infection
- PMID: 29192235
- PMCID: PMC5709414
- DOI: 10.1038/s41467-017-01693-z
Multiplex single-cell visualization of nucleic acids and protein during HIV infection
Abstract
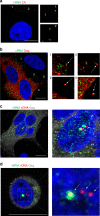
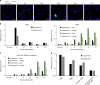
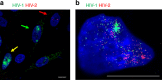
Technical limitations in simultaneous microscopic visualization of RNA, DNA, and proteins of HIV have curtailed progress in this field. To address this need we develop a microscopy approach, multiplex immunofluorescent cell-based detection of DNA, RNA and Protein (MICDDRP), which is based on branched DNA in situ hybridization technology. MICDDRP enables simultaneous single-cell visualization of HIV (a) spliced and unspliced RNA, (b) cytoplasmic and nuclear DNA, and (c) Gag. We use MICDDRP to visualize incoming capsid cores containing RNA and/or nascent DNA and follow reverse transcription kinetics. We also report transcriptional "bursts" of nascent RNA from integrated proviral DNA, and concomitant HIV-1, HIV-2 transcription in co-infected cells. MICDDRP can be used to simultaneously detect multiple viral nucleic acid intermediates, characterize the effects of host factors or drugs on steps of the HIV life cycle, or its reactivation from the latent state, thus facilitating the development of antivirals and latency reactivating agents.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Similar articles
-
Single-cell Multiplexed Fluorescence Imaging to Visualize Viral Nucleic Acids and Proteins and Monitor HIV, HTLV, HBV, HCV, Zika Virus, and Influenza Infection.J Vis Exp. 2020 Oct 29;(164):10.3791/61843. doi: 10.3791/61843. J Vis Exp. 2020. PMID: 33191939 Free PMC article.
-
Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells.Viruses. 2018 Sep 30;10(10):534. doi: 10.3390/v10100534. Viruses. 2018. PMID: 30274333 Free PMC article.
-
Human immunodeficiency virus type 1 nucleocapsid protein nuclear localization mediates early viral mRNA expression.J Virol. 2002 Oct;76(20):10444-54. doi: 10.1128/jvi.76.20.10444-10454.2002. J Virol. 2002. PMID: 12239321 Free PMC article.
-
Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism.Prog Nucleic Acid Res Mol Biol. 2005;80:217-86. doi: 10.1016/S0079-6603(05)80006-6. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164976 Review. No abstract available.
-
Distribution and fate of HIV-1 unintegrated DNA species: a comprehensive update.AIDS Res Ther. 2017 Feb 16;14(1):9. doi: 10.1186/s12981-016-0127-6. AIDS Res Ther. 2017. PMID: 28209198 Free PMC article. Review.
Cited by
-
Remodeling of the Core Leads HIV-1 Preintegration Complex into the Nucleus of Human Lymphocytes.J Virol. 2020 May 18;94(11):e00135-20. doi: 10.1128/JVI.00135-20. Print 2020 May 18. J Virol. 2020. PMID: 32238582 Free PMC article.
-
HIV-2 inhibits HIV-1 gene expression via two independent mechanisms during cellular co-infection.J Virol. 2023 Dec 21;97(12):e0187022. doi: 10.1128/jvi.01870-22. Epub 2023 Nov 22. J Virol. 2023. PMID: 37991365 Free PMC article.
-
Characterization of Inducible Transcription and Translation-Competent HIV-1 Using the RNAscope ISH Technology at a Single-Cell Resolution.Front Microbiol. 2018 Oct 2;9:2358. doi: 10.3389/fmicb.2018.02358. eCollection 2018. Front Microbiol. 2018. PMID: 30333813 Free PMC article.
-
Deep Parallel Characterization of AAV Tropism and AAV-Mediated Transcriptional Changes via Single-Cell RNA Sequencing.Front Immunol. 2021 Oct 21;12:730825. doi: 10.3389/fimmu.2021.730825. eCollection 2021. Front Immunol. 2021. PMID: 34759919 Free PMC article.
-
HIV-1 capsid stability and reverse transcription are finely balanced to minimize sensing of reverse transcription products via the cGAS-STING pathway.mBio. 2024 May 8;15(5):e0034824. doi: 10.1128/mbio.00348-24. Epub 2024 Mar 26. mBio. 2024. PMID: 38530034 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

