ClinVar: improving access to variant interpretations and supporting evidence
- PMID: 29165669
- PMCID: PMC5753237
- DOI: 10.1093/nar/gkx1153
ClinVar: improving access to variant interpretations and supporting evidence
Abstract
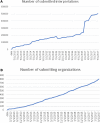
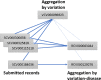
ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) is a freely available, public archive of human genetic variants and interpretations of their significance to disease, maintained at the National Institutes of Health. Interpretations of the clinical significance of variants are submitted by clinical testing laboratories, research laboratories, expert panels and other groups. ClinVar aggregates data by variant-disease pairs, and by variant (or set of variants). Data aggregated by variant are accessible on the website, in an improved set of variant call format files and as a new comprehensive XML report. ClinVar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing. Submissions may come from clinical providers providing their own interpretation of the variant ('provider interpretation') or from groups such as patient registries that primarily provide phenotypic information from patients ('phenotyping only'). ClinVar continues to make improvements to its search and retrieval functions. Several new fields are now indexed for more precise searching, and filters allow the user to narrow down a large set of search results.
Published by Oxford University Press on behalf of Nucleic Acids Research 2017.
Figures
Similar articles
-
ClinVar: improvements to accessing data.Nucleic Acids Res. 2020 Jan 8;48(D1):D835-D844. doi: 10.1093/nar/gkz972. Nucleic Acids Res. 2020. PMID: 31777943 Free PMC article.
-
ClinVar: public archive of interpretations of clinically relevant variants.Nucleic Acids Res. 2016 Jan 4;44(D1):D862-8. doi: 10.1093/nar/gkv1222. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26582918 Free PMC article.
-
ClinVar: public archive of relationships among sequence variation and human phenotype.Nucleic Acids Res. 2014 Jan;42(Database issue):D980-5. doi: 10.1093/nar/gkt1113. Epub 2013 Nov 14. Nucleic Acids Res. 2014. PMID: 24234437 Free PMC article.
-
Somatic cancer variant curation and harmonization through consensus minimum variant level data.Genome Med. 2016 Nov 4;8(1):117. doi: 10.1186/s13073-016-0367-z. Genome Med. 2016. PMID: 27814769 Free PMC article.
-
Assessment of an automated approach for variant interpretation in screening for monogenic disorders: A single-center study.Mol Genet Genomic Med. 2022 Dec;10(12):e2085. doi: 10.1002/mgg3.2085. Epub 2022 Nov 5. Mol Genet Genomic Med. 2022. PMID: 36333997 Free PMC article. Review.
Cited by
-
Extension of the phenotypic spectrum of GLE1-related disorders to a mild congenital form resembling congenital myopathy.Mol Genet Genomic Med. 2020 Aug;8(8):e1277. doi: 10.1002/mgg3.1277. Epub 2020 Jun 14. Mol Genet Genomic Med. 2020. PMID: 32537934 Free PMC article.
-
Sex-specific survival gene mutations are discovered as clinical predictors of clear cell renal cell carcinoma.Sci Rep. 2024 Jul 9;14(1):15800. doi: 10.1038/s41598-024-66525-9. Sci Rep. 2024. PMID: 38982123 Free PMC article.
-
Genomic study of nonsyndromic hearing loss in unaffected individuals: Frequency of pathogenic and likely pathogenic variants in a Brazilian cohort of 2,097 genomes.Front Genet. 2022 Aug 30;13:921324. doi: 10.3389/fgene.2022.921324. eCollection 2022. Front Genet. 2022. PMID: 36147510 Free PMC article.
-
Dynamic incorporation of multiple in silico functional annotations empowers rare variant association analysis of large whole-genome sequencing studies at scale.Nat Genet. 2020 Sep;52(9):969-983. doi: 10.1038/s41588-020-0676-4. Epub 2020 Aug 24. Nat Genet. 2020. PMID: 32839606 Free PMC article.
-
Impact of genome build on RNA-seq interpretation and diagnostics.medRxiv [Preprint]. 2024 Jan 12:2024.01.11.24301165. doi: 10.1101/2024.01.11.24301165. medRxiv. 2024. Update in: Am J Hum Genet. 2024 Jul 11;111(7):1282-1300. doi: 10.1016/j.ajhg.2024.05.005 PMID: 38260490 Free PMC article. Updated. Preprint.
References
-
- den Dunnen J.T., Antonarakis S.E.. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000; 15:7–12. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

