Members of Bitter Taste Receptor Cluster Tas2r143/Tas2r135/Tas2r126 Are Expressed in the Epithelium of Murine Airways and Other Non-gustatory Tissues
- PMID: 29163195
- PMCID: PMC5670347
- DOI: 10.3389/fphys.2017.00849
Members of Bitter Taste Receptor Cluster Tas2r143/Tas2r135/Tas2r126 Are Expressed in the Epithelium of Murine Airways and Other Non-gustatory Tissues
Abstract
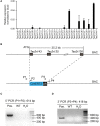
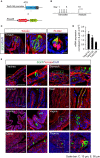
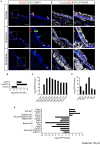
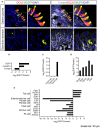
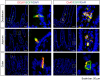
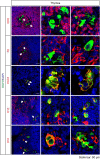
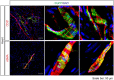
The mouse bitter taste receptors Tas2r143, Tas2r135, and Tas2r126 are encoded by genes that cluster on chromosome 6 and have been suggested to be expressed under common regulatory elements. Previous studies indicated that the Tas2r143/Tas2r135/Tas2r126 cluster is expressed in the heart, but other organs had not been systematically analyzed. In order to investigate the expression of this bitter taste receptor gene cluster in non-gustatory tissues, we generated a BAC (bacterial artificial chromosome) based transgenic mouse line, expressing CreERT2 under the control of the Tas2r143 promoter. After crossing this line with a mouse line expressing EGFP after Cre-mediated recombination, we were able to validate the Tas2r143-CreERT2 transgenic mouse line and monitor the expression of Tas2r143. EGFP-positive cells, indicating expression of members of the cluster, were found in about 47% of taste buds, and could also be found in several other organs. A population of EGFP-positive cells was identified in thymic epithelial cells, in the lamina propria of the intestine and in vascular smooth muscle cells of cardiac blood vessels. EGFP-positive cells were also identified in the epithelium of organs readily exposed to pathogens including lower airways, the gastrointestinal tract, urethra, vagina, and cervix. With respect to the function of cells expressing this bitter taste receptor cluster, RNA-seq analysis in EGFP-positive cells isolated from the epithelium of trachea and stomach showed expression of genes related to innate immunity. These data further support the concept that bitter taste receptors serve functions outside the gustatory system.
Keywords: Tas2r126; Tas2r135; Tas2r143; chemosensory cells; tuft cells.
Figures

Similar articles
-
Genetic deletion of the Tas2r143/Tas2r135/Tas2r126 cluster reveals that TAS2Rs may not mediate bitter tastant-induced bronchodilation.J Cell Physiol. 2021 Sep;236(9):6407-6423. doi: 10.1002/jcp.30315. Epub 2021 Feb 8. J Cell Physiol. 2021. PMID: 33559206 Free PMC article.
-
Bitter tastants relax the mouse gallbladder smooth muscle independent of signaling through tuft cells and bitter taste receptors.Sci Rep. 2024 Aug 8;14(1):18447. doi: 10.1038/s41598-024-69287-6. Sci Rep. 2024. PMID: 39117690 Free PMC article.
-
Distinct Cell Types With the Bitter Receptor Tas2r126 in Different Compartments of the Stomach.Front Physiol. 2020 Feb 7;11:32. doi: 10.3389/fphys.2020.00032. eCollection 2020. Front Physiol. 2020. PMID: 32116750 Free PMC article.
-
Oral and extraoral bitter taste receptors.Results Probl Cell Differ. 2010;52:87-99. doi: 10.1007/978-3-642-14426-4_8. Results Probl Cell Differ. 2010. PMID: 20865374 Review.
-
Building sensory receptors on the tongue.J Neurocytol. 2004 Dec;33(6):631-46. doi: 10.1007/s11068-005-3332-0. Epub 2005 Oct 11. J Neurocytol. 2004. PMID: 16217619 Review.
Cited by
-
Phenanthroline relaxes uterine contractions induced by diverse contractile agents by decreasing cytosolic calcium concentration.Eur J Pharmacol. 2024 Apr 5;968:176343. doi: 10.1016/j.ejphar.2024.176343. Epub 2024 Jan 26. Eur J Pharmacol. 2024. PMID: 38281680
-
Genetic deletion of the Tas2r143/Tas2r135/Tas2r126 cluster reveals that TAS2Rs may not mediate bitter tastant-induced bronchodilation.J Cell Physiol. 2021 Sep;236(9):6407-6423. doi: 10.1002/jcp.30315. Epub 2021 Feb 8. J Cell Physiol. 2021. PMID: 33559206 Free PMC article.
-
Bitter tastants relax the mouse gallbladder smooth muscle independent of signaling through tuft cells and bitter taste receptors.Sci Rep. 2024 Aug 8;14(1):18447. doi: 10.1038/s41598-024-69287-6. Sci Rep. 2024. PMID: 39117690 Free PMC article.
-
Bitter Taste Receptors (T2Rs) are Sentinels that Coordinate Metabolic and Immunological Defense Responses.Curr Opin Physiol. 2021 Apr;20:70-76. doi: 10.1016/j.cophys.2021.01.006. Epub 2021 Jan 12. Curr Opin Physiol. 2021. PMID: 33738371 Free PMC article.
-
Genetic mutation of Tas2r104/Tas2r105/Tas2r114 cluster leads to a loss of taste perception to denatonium benzoate and cucurbitacin B.Animal Model Exp Med. 2024 Jun;7(3):324-336. doi: 10.1002/ame2.12357. Epub 2023 Dec 28. Animal Model Exp Med. 2024. PMID: 38155461 Free PMC article.
References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at http://www.bioinformatics.babraham.ac.uk/projects/fastqc
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

