Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication
- PMID: 29162711
- PMCID: PMC5698553
- DOI: 10.1128/mBio.01658-17
Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication
Abstract
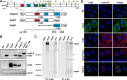
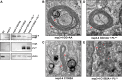
Betacoronaviruses, such as Middle East respiratory syndrome coronavirus (MERS-CoV), are important pathogens causing potentially lethal infections in humans and animals. Coronavirus RNA synthesis is thought to be associated with replication organelles (ROs) consisting of modified endoplasmic reticulum (ER) membranes. These are transformed into double-membrane vesicles (DMVs) containing viral double-stranded RNA and into other membranous elements such as convoluted membranes, together forming a reticulovesicular network. Previous evidence suggested that the nonstructural proteins (nsp's) 3, 4, and 6 of the severe acute respiratory syndrome coronavirus (SARS-CoV), which contain transmembrane domains, would all be required for DMV formation. We have now expressed MERS-CoV replicase self-cleaving polyprotein fragments encompassing nsp3-4 or nsp3-6, as well as coexpressed nsp3 and nsp4 of either MERS-CoV or SARS-CoV, to characterize the membrane structures induced. Using electron tomography, we demonstrate that for both MERS-CoV and SARS-CoV coexpression of nsp3 and nsp4 is required and sufficient to induce DMVs. Coexpression of MERS-CoV nsp3 and nsp4 either as individual proteins or as a self-cleaving nsp3-4 precursor resulted in very similar DMVs, and in both setups we observed proliferation of zippered ER that appeared to wrap into nascent DMVs. Moreover, when inactivating nsp3-4 polyprotein cleavage by mutagenesis, we established that cleavage of the nsp3/nsp4 junction is essential for MERS-CoV DMV formation. Addition of the third MERS-CoV transmembrane protein, nsp6, did not noticeably affect DMV formation. These findings provide important insight into the biogenesis of coronavirus DMVs, establish strong similarities with other nidoviruses (specifically, the arteriviruses), and highlight possible general principles in viral DMV formation.IMPORTANCE The RNA replication of positive stranded RNA viruses of eukaryotes is thought to take place at cytoplasmic membranous replication organelles (ROs). Double-membrane vesicles are a prominent type of viral ROs. They are induced by coronaviruses, such as SARS-CoV and MERS-CoV, as well as by a number of other important pathogens, yet little is known about their biogenesis. In this study, we explored the viral protein requirements for the formation of MERS-CoV- and SARS-CoV-induced DMVs and established that coexpression of two of the three transmembrane subunits of the coronavirus replicase polyprotein, nonstructural proteins (nsp's) 3 and 4, is required and sufficient to induce DMV formation. Moreover, release of nsp3 and nsp4 from the polyprotein by proteolytic maturation is essential for this process. These findings provide a strong basis for further research on the biogenesis and functionality of coronavirus ROs and may point to more general principles of viral DMV formation.
Keywords: convoluted membranes; electron tomography; membrane structure; nidoviruses; nonstructural proteins; replication complex; replication organelle biogenesis; replication structures; viral factory; viral protein.
Copyright © 2017 Oudshoorn et al.
Figures




Similar articles
-
Oligomeric assembly of the C-terminal and transmembrane region of SARS-CoV-2 nsp3.J Virol. 2024 Apr 16;98(4):e0157523. doi: 10.1128/jvi.01575-23. Epub 2024 Mar 14. J Virol. 2024. PMID: 38483167 Free PMC article.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
-
Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications.J Virol. 2015 Feb;89(4):2080-9. doi: 10.1128/JVI.02776-14. Epub 2014 Dec 3. J Virol. 2015. PMID: 25473044 Free PMC article.
-
NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication.mBio. 2015 Jul 7;6(4):e00759. doi: 10.1128/mBio.00759-15. mBio. 2015. PMID: 26152585 Free PMC article.
-
The double-membrane vesicle (DMV): a virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses.Cell Mol Life Sci. 2022 Jul 16;79(8):425. doi: 10.1007/s00018-022-04469-x. Cell Mol Life Sci. 2022. PMID: 35841484 Free PMC article. Review.
Cited by
-
Positive-strand RNA virus replication organelles at a glance.J Cell Sci. 2024 Sep 1;137(17):jcs262164. doi: 10.1242/jcs.262164. Epub 2024 Sep 10. J Cell Sci. 2024. PMID: 39254430 Free PMC article. Review.
-
Molecular architecture of coronavirus double-membrane vesicle pore complex.Nature. 2024 Sep;633(8028):224-231. doi: 10.1038/s41586-024-07817-y. Epub 2024 Aug 14. Nature. 2024. PMID: 39143215 Free PMC article.
-
ACADM inhibits AMPK activation to modulate PEDV-induced lipophagy and β-oxidation for impairing viral replication.J Biol Chem. 2024 Aug;300(8):107549. doi: 10.1016/j.jbc.2024.107549. Epub 2024 Jul 11. J Biol Chem. 2024. PMID: 39002673 Free PMC article.
-
The glycoprotein quality control factor Malectin promotes coronavirus replication and viral protein biogenesis.bioRxiv [Preprint]. 2024 Jul 16:2024.06.02.597051. doi: 10.1101/2024.06.02.597051. bioRxiv. 2024. PMID: 38895409 Free PMC article. Preprint.
-
Nanoscale cellular organization of viral RNA and proteins in SARS-CoV-2 replication organelles.Nat Commun. 2024 May 31;15(1):4644. doi: 10.1038/s41467-024-48991-x. Nat Commun. 2024. PMID: 38821943 Free PMC article.
References
-
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguière AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi:10.1056/NEJMoa030747. - DOI - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi:10.1056/NEJMoa030781. - DOI - PubMed
-
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473-12. doi:10.1128/mBio.00473-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
