Multi-platform 'Omics Analysis of Human Ebola Virus Disease Pathogenesis
- PMID: 29154144
- PMCID: PMC5730472
- DOI: 10.1016/j.chom.2017.10.011
Multi-platform 'Omics Analysis of Human Ebola Virus Disease Pathogenesis
Abstract
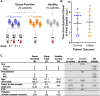
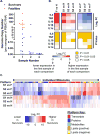
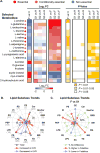
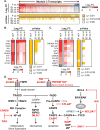
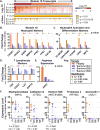
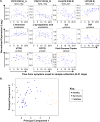
The pathogenesis of human Ebola virus disease (EVD) is complex. EVD is characterized by high levels of virus replication and dissemination, dysregulated immune responses, extensive virus- and host-mediated tissue damage, and disordered coagulation. To clarify how host responses contribute to EVD pathophysiology, we performed multi-platform 'omics analysis of peripheral blood mononuclear cells and plasma from EVD patients. Our results indicate that EVD molecular signatures overlap with those of sepsis, imply that pancreatic enzymes contribute to tissue damage in fatal EVD, and suggest that Ebola virus infection may induce aberrant neutrophils whose activity could explain hallmarks of fatal EVD. Moreover, integrated biomarker prediction identified putative biomarkers from different data platforms that differentiated survivors and fatalities early after infection. This work reveals insight into EVD pathogenesis, suggests an effective approach for biomarker identification, and provides an important community resource for further analysis of human EVD severity.
Keywords: Ebola virus; biomarker; human; inflammation; neutrophils; omics; pancreatitis; pathogenesis; sepsis; systems biology.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors do not have any conflicts of interest to declare.
Figures

Similar articles
-
Plasma Proteomic Analysis Distinguishes Severity Outcomes of Human Ebola Virus Disease.mBio. 2022 Jun 28;13(3):e0056722. doi: 10.1128/mbio.00567-22. Epub 2022 Apr 21. mBio. 2022. PMID: 35446128 Free PMC article.
-
Clinical, virological, and biological parameters associated with outcomes of Ebola virus infection in Macenta, Guinea.JCI Insight. 2017 Mar 23;2(6):e88864. doi: 10.1172/jci.insight.88864. JCI Insight. 2017. PMID: 28352651 Free PMC article.
-
Avatar Mice Underscore the Role of the T Cell-Dendritic Cell Crosstalk in Ebola Virus Disease and Reveal Mechanisms of Protection in Survivors.J Virol. 2022 Sep 28;96(18):e0057422. doi: 10.1128/jvi.00574-22. Epub 2022 Sep 8. J Virol. 2022. PMID: 36073921 Free PMC article.
-
Clinical Manifestations and Pathogenesis of Uveitis in Ebola Virus Disease Survivors.Ocul Immunol Inflamm. 2018;26(7):1128-1134. doi: 10.1080/09273948.2018.1484493. Epub 2018 Jul 11. Ocul Immunol Inflamm. 2018. PMID: 29993303 Review.
-
[Ebola Virus Disease (Ebola Hemorrhagic Fever)].Rinsho Byori. 2016 Sep;64(9):1025-1032. Rinsho Byori. 2016. PMID: 30609455 Review. Japanese.
Cited by
-
Serum lipidome reveals lipid metabolic dysregulation in severe fever with thrombocytopenia syndrome.BMC Med. 2024 Oct 14;22(1):458. doi: 10.1186/s12916-024-03672-w. BMC Med. 2024. PMID: 39396989 Free PMC article.
-
Plasma Metabolomic Profiles in Recovered COVID-19 Patients without Previous Underlying Diseases 3 Months After Discharge.J Inflamm Res. 2021 Sep 7;14:4485-4501. doi: 10.2147/JIR.S325853. eCollection 2021. J Inflamm Res. 2021. PMID: 34522117 Free PMC article.
-
Characterization of Ebola Virus Mucosal Challenge Routes in Cynomolgus Macaques.J Virol. 2023 May 31;97(5):e0188822. doi: 10.1128/jvi.01888-22. Epub 2023 Mar 28. J Virol. 2023. PMID: 36975793 Free PMC article.
-
Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2.Int J Mol Sci. 2020 Nov 16;21(22):8623. doi: 10.3390/ijms21228623. Int J Mol Sci. 2020. PMID: 33207699 Free PMC article.
-
Analysis of an Ebola virus disease survivor whose host and viral markers were predictive of death indicates the effectiveness of medical countermeasures and supportive care.Genome Med. 2021 Jan 11;13(1):5. doi: 10.1186/s13073-020-00811-9. Genome Med. 2021. PMID: 33430949 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

