INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division
- PMID: 29142021
- PMCID: PMC5748994
- DOI: 10.1083/jcb.201709111
INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division
Abstract
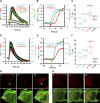
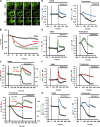
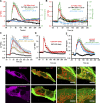
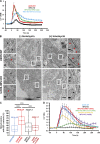
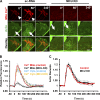
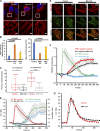
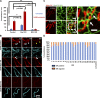
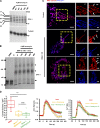
Mitochondrial division requires division of both the inner and outer mitochondrial membranes (IMM and OMM, respectively). Interaction with endoplasmic reticulum (ER) promotes OMM division by recruitment of the dynamin Drp1, but effects on IMM division are not well characterized. We previously showed that actin polymerization through ER-bound inverted formin 2 (INF2) stimulates Drp1 recruitment in mammalian cells. Here, we show that INF2-mediated actin polymerization stimulates a second mitochondrial response independent of Drp1: a rise in mitochondrial matrix calcium through the mitochondrial calcium uniporter. ER stores supply the increased mitochondrial calcium, and the role of actin is to increase ER-mitochondria contact. Myosin IIA is also required for this mitochondrial calcium increase. Elevated mitochondrial calcium in turn activates IMM constriction in a Drp1-independent manner. IMM constriction requires electron transport chain activity. IMM division precedes OMM division. These results demonstrate that actin polymerization independently stimulates the dynamics of both membranes during mitochondrial division: IMM through increased matrix calcium, and OMM through Drp1 recruitment.
© 2018 Chakrabarti et al.
Figures

Comment in
-
Organelles: The Emerging Signalling Chart of Mitochondrial Dynamics.Curr Biol. 2018 Jan 22;28(2):R73-R75. doi: 10.1016/j.cub.2017.11.040. Curr Biol. 2018. PMID: 29374448
Similar articles
-
An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2.Science. 2013 Jan 25;339(6118):464-7. doi: 10.1126/science.1228360. Science. 2013. PMID: 23349293 Free PMC article.
-
Receptor-mediated Drp1 oligomerization on endoplasmic reticulum.J Cell Biol. 2017 Dec 4;216(12):4123-4139. doi: 10.1083/jcb.201610057. Epub 2017 Nov 20. J Cell Biol. 2017. PMID: 29158231 Free PMC article.
-
Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division.Nat Commun. 2017 Jun 9;8:15754. doi: 10.1038/ncomms15754. Nat Commun. 2017. PMID: 28598422 Free PMC article.
-
The Complex Dance of Organelles during Mitochondrial Division.Trends Cell Biol. 2021 Apr;31(4):241-253. doi: 10.1016/j.tcb.2020.12.005. Epub 2021 Jan 11. Trends Cell Biol. 2021. PMID: 33446409 Review.
-
It takes two to tango: The essential role of ER-mitochondrial contact sites in mitochondrial dynamics.Int J Biochem Cell Biol. 2021 Dec;141:106101. doi: 10.1016/j.biocel.2021.106101. Epub 2021 Oct 23. Int J Biochem Cell Biol. 2021. PMID: 34695569 Review.
Cited by
-
Mitochondrial mechanotransduction through MIEF1 coordinates the nuclear response to forces.Nat Cell Biol. 2024 Dec;26(12):2046-2060. doi: 10.1038/s41556-024-01527-3. Epub 2024 Oct 21. Nat Cell Biol. 2024. PMID: 39433949 Free PMC article.
-
Mitochondrial dysfunction triggers actin polymerization necessary for rapid glycolytic activation.J Cell Biol. 2022 Nov 7;221(11):e202201160. doi: 10.1083/jcb.202201160. Epub 2022 Sep 14. J Cell Biol. 2022. PMID: 36102863 Free PMC article.
-
Therapeutic Effects of Mesenchymal Stromal Cells Require Mitochondrial Transfer and Quality Control.Int J Mol Sci. 2023 Oct 31;24(21):15788. doi: 10.3390/ijms242115788. Int J Mol Sci. 2023. PMID: 37958771 Free PMC article. Review.
-
Mitochondria-Endoplasmic Reticulum Contacts: The Promising Regulators in Diabetic Cardiomyopathy.Oxid Med Cell Longev. 2022 Apr 11;2022:2531458. doi: 10.1155/2022/2531458. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35450404 Free PMC article. Review.
-
Mitochondria-actin cytoskeleton crosstalk in cell migration.J Cell Physiol. 2022 May;237(5):2387-2403. doi: 10.1002/jcp.30729. Epub 2022 Mar 27. J Cell Physiol. 2022. PMID: 35342955 Free PMC article. Review.
References
-
- Baffy G., Miyashita T., Williamson J.R., and Reed J.C.. 1993. Apoptosis induced by withdrawal of interleukin-3 (IL-3) from an IL-3-dependent hematopoietic cell line is associated with repartitioning of intracellular calcium and is blocked by enforced Bcl-2 oncoprotein production. J. Biol. Chem. 268:6511–6519. - PubMed
-
- Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L., et al. . 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 476:341–345. 10.1038/nature10234 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

