Cellular v-ATPase is required for virion assembly compartment formation in human cytomegalovirus infection
- PMID: 29093211
- PMCID: PMC5717334
- DOI: 10.1098/rsob.160298
Cellular v-ATPase is required for virion assembly compartment formation in human cytomegalovirus infection
Abstract
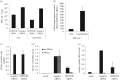
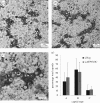
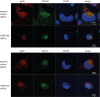
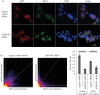
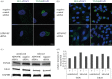
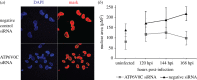
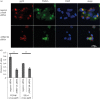
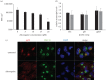
Successful generation of virions from infected cells is a complex process requiring orchestrated regulation of host and viral genes. Cells infected with human cytomegalovirus (HCMV) undergo a dramatic reorganization of membrane organelles resulting in the formation of the virion assembly compartment, a process that is not fully understood. Here we show that acidification of vacuoles by the cellular v-ATPase is a crucial step in the formation of the virion assembly compartment and disruption of acidification results in mis-localization of virion components and a profound reduction in infectious virus levels. In addition, knockdown of ATP6V0C blocks the increase in nuclear size, normally associated with HCMV infection. Inhibition of the v-ATPase does not affect intracellular levels of viral DNA synthesis or gene expression, consistent with a defect in assembly and egress. These studies identify a novel host factor involved in virion production and a potential target for antiviral therapy.
Keywords: assembly and egress; herpesvirus; host virus interaction; human cytomegalovirus; v-ATPase.
© 2017 The Authors.
Conflict of interest statement
The authors have no competing interests.
Figures

Similar articles
-
Human Cytomegalovirus Envelope Protein gpUL132 Regulates Infectious Virus Production through Formation of the Viral Assembly Compartment.mBio. 2020 Sep 29;11(5):e02044-20. doi: 10.1128/mBio.02044-20. mBio. 2020. PMID: 32994323 Free PMC article.
-
Phosphorylation of Golgi Peripheral Membrane Protein Grasp65 Is an Integral Step in the Formation of the Human Cytomegalovirus Cytoplasmic Assembly Compartment.mBio. 2016 Oct 4;7(5):e01554-16. doi: 10.1128/mBio.01554-16. mBio. 2016. PMID: 27703074 Free PMC article.
-
Human Cytomegalovirus Hijacks WD Repeat Domain 11 for Virion Assembly Compartment Formation and Virion Morphogenesis.J Virol. 2022 Mar 9;96(5):e0182721. doi: 10.1128/JVI.01827-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35020472 Free PMC article.
-
Betaherpesvirus assembly and egress: Recent advances illuminate the path.Adv Virus Res. 2020;108:337-392. doi: 10.1016/bs.aivir.2020.09.003. Epub 2020 Oct 7. Adv Virus Res. 2020. PMID: 33837722 Review.
-
Host Cell Signatures of the Envelopment Site within Beta-Herpes Virions.Int J Mol Sci. 2022 Sep 1;23(17):9994. doi: 10.3390/ijms23179994. Int J Mol Sci. 2022. PMID: 36077391 Free PMC article. Review.
Cited by
-
Systems Virology and Human Cytomegalovirus: Using High Throughput Approaches to Identify Novel Host-Virus Interactions During Lytic Infection.Front Cell Infect Microbiol. 2020 Jun 10;10:280. doi: 10.3389/fcimb.2020.00280. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32587832 Free PMC article. Review.
-
A Comprehensive Proteomics Analysis of the JC Virus (JCV) Large and Small Tumor Antigen Interacting Proteins: Large T Primarily Targets the Host Protein Complexes with V-ATPase and Ubiquitin Ligase Activities While Small t Mostly Associates with Those Having Phosphatase and Chromatin-Remodeling Functions.Viruses. 2020 Oct 20;12(10):1192. doi: 10.3390/v12101192. Viruses. 2020. PMID: 33092197 Free PMC article.
-
Virus-host protein interactions as footprints of human cytomegalovirus replication.Curr Opin Virol. 2022 Feb;52:135-147. doi: 10.1016/j.coviro.2021.11.016. Epub 2021 Dec 16. Curr Opin Virol. 2022. PMID: 34923282 Free PMC article. Review.
-
Organelle dynamics and viral infections: at cross roads.Microbes Infect. 2019 Jan-Feb;21(1):20-32. doi: 10.1016/j.micinf.2018.06.002. Epub 2018 Jun 25. Microbes Infect. 2019. PMID: 29953921 Free PMC article. Review.
-
V-ATPases and osteoclasts: ambiguous future of V-ATPases inhibitors in osteoporosis.Theranostics. 2018 Oct 26;8(19):5379-5399. doi: 10.7150/thno.28391. eCollection 2018. Theranostics. 2018. PMID: 30555553 Free PMC article. Review.
References
-
- Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143, 222–234. (doi:10.1016/j.virusres.2009.03.018) - DOI - PubMed
-
- Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394. (doi:10.1038/nrmicro2559) - DOI - PubMed
-
- Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74, 975–986. (doi:10.1128/jvi.74.2.975-986.2000) - DOI - PMC - PubMed
-
- Buchkovich NJ, Maguire TG, Alwine JC. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J. Virol. 84, 7005–7017. (doi:10.1128/JVI.00719-10) - DOI - PMC - PubMed
-
- Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81, 11 861–11 869. (doi:10.1128/jvi.01077-07) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

